Abstract
Tumor-derived microvesicles (TMVs) comprise a class of extracellular vesicles released from tumor cells that are now understood to facilitate communication between the tumor and the surrounding microenvironment. Despite their significance, the regulatory mechanisms governing the trafficking of bioactive cargos to TMVs at the cell surface remain poorly defined. Here we describe a molecular pathway for the delivery of microRNA (miRNA) cargo to nascent TMVs involving the dissociation of a pre-miRNA/Exportin-5 complex from Ran-GTP following nuclear export, and its subsequent transfer to a cytoplasmic shuttle comprised of ARF6-GTP and GRP1. As such, ARF6 activation increases pre-miRNA cargo contained within TMVs via a process that requires casein kinase 2-mediated phosphorylation of Ran-GAP1. Further, TMVs were found to contain pre-miRNA processing machinery including Dicer and Argonaute 2, which allow for cell-free pre-miRNA processing within shed vesicles. These findings offer cellular targets to block the loading and processing of pre-miRNAs within TMVs.
Introduction:
The regulated shedding of extracellular vesicles (EVs) is now understood to be an important means of intercellular communication1-3. Among these, microvesicles released from tumor cells, often called tumor-derived microvesicles (TMVs), have emerged as crucial participants in tumor pathogenesis, metastasis, and communication4-7. Unlike another class of EVs, tumor-derived exosomes, TMVs are generated through direct budding from plasma membrane into the extracellular space where they can interact with and influence recipient cells within the local tumor microenvironment or at distal sites2-4,8. Direct release into the extracellular space, allows for TMV detection and isolation from peripheral bodily fluids5,9.
TMV biogenesis is tightly regulated with cellular control exerted over the inclusion of specific cargo, and fission events that release the vesicle from the originating cell1,5. TMV shedding it’s now known is, at least in part, regulated by signaling pathways governed by the activation of small GTPases including ADP-ribosylation factor 6 (ARF6), RhoA, and Rab22A3,4,6,8,10. TMV budding converges on the activation of intrinsic contractile machinery in order to facilitate the fission and release of mature TMVs6,8. While some cargo originates from recycling endosomal membranes, certain other endosomal components are excluded from shed TMVs8, highlighting the selective recruitment of TMV cargo. For example, association between MT1-MMP and the vSNARE VAMP-3, was shown to direct trafficking of protease cargo through the endosomal system to sites of TMV formation5. Additionally, studies have shown that the Arrestin 1 domain–containing protein 1 (ARRDC1), recruits the ESCRT-I subunit, TSG101, to the plasma membrane resulting in the release of small microvesicles containing TSG101, ARRDC1, and other cellular proteins11-13. Further study of regulators of TMV cargo trafficking, delivery, and inclusion/exclusion is necessary to advance our understanding of TMV biogenesis.
Many functional consequences of TMV release stem from their diverse bioactive cargo, which includes proteases, cell-surface receptors, nucleic acids, and active lipids4,5,14-16. Horizontal transfer of bioactive cargoes represents a mechanism through which these vesicles can alter recipient cells14,16-19. Among the nucleic acids capable of altering recipient cell gene expression, small RNAs, especially miRNAs are increasingly important. The deregulation of miRNAs has been documented in multiple cancers where they’re known to target tumor suppressors and related proteins resulting in poor patient outcomes20-24.
MicroRNAs represent the dominant class of small RNAs evolved to suppress unwanted genetic material and transcripts in somatic tissues. Their processing begins with transcription of primary miRNA (pri-miRNA) sequences containing one or more hairpin structures and a terminal loop. Prior to nuclear export, pri-miRNAs are processed to pre-miRNA by the microprocessor complex containing DGCR8 and Drosha. Pre-miRNA is exported from the nucleus by the Importin β-like nuclear export receptor Exportin-5 together with RanGTP25,26 where it’s cleaved into a ~22 nt miRNA duplex by Dicer before one of the strands is loaded into the RNA-induced silencing complex (RISC) together with a member of the Argonaute family of proteins27.
In general, RNA cargo contained within EVs reflects the levels and types of cytoplasmic content; is determined by the mode of EV biogenesis; and type and physiologic state of the releasing cell1,28. Since the identification of miRNAs within EVs, various mechanisms have been proposed for their selection into exosomes. RNA-binding proteins (RBPs), Y-box protein 1 (YBX1) and sumoylated hnRNPA2B1, are reported to bind miRNAs at specific sequence motifs and mediate their trafficking to exosomes29,30. Over-expression of nSMase2 has been shown to increase exosomal miRNAs31,32. Argonaute-2, a protein associated with the RISC machinery is thought to control the loading of miRNA in part through KRAS-dependent association of P-bodies with the cytoplasmic membrane of multi-vesicular bodies33-35. Furthermore, the presence of pre-miRNA in exosomes, along with CD43-mediated Dicer accumulation was previously reported to allow for pre-miRNA processing within exosomes14.
The mechanisms of pre-miRNA sorting and incorporation into TMVs is far less defined. Here we report that interaction between ARF6-GTP and the importin β-like nuclear export receptor, Exportin-5, directs trafficking of a pre-miRNA complex to sites of TMV biogenesis for inclusion as TMV cargo. This involves a hand off of pre-miRNA together with Exportin-5 from a RanGTP nuclear export complex, to an ARF-GEF scaffold that is essential for the ARF6-GTP-dependent delivery to TMVs. Disrupting the ability of Exportin-5 to interact with ARF6 results not only in the release of TMVs lacking Exportin-5, but a subsequent decrease in TMV miRNA cargo.
Results:
Invasive Tumor Cells shed TMVs containing bioactive cargo, including miRNAs.
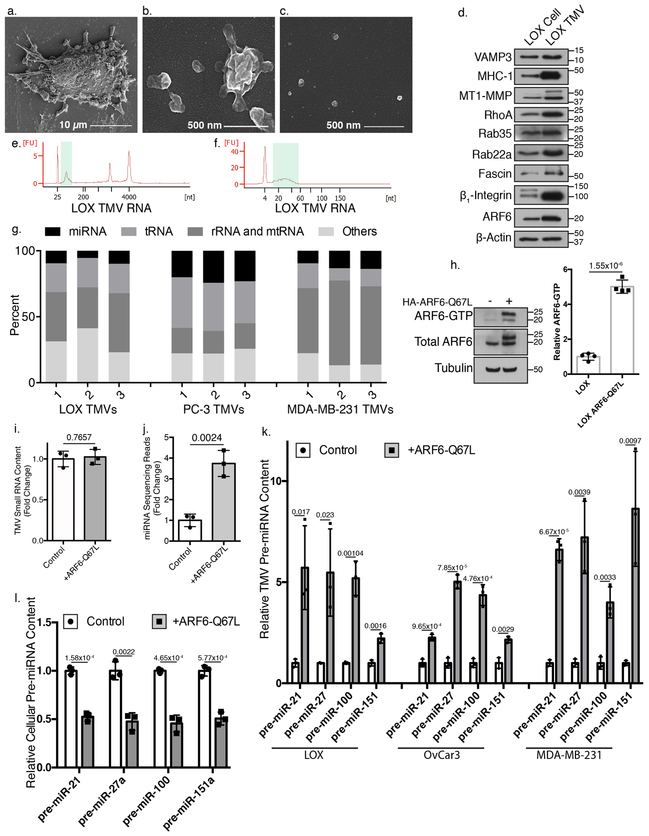
Invasive tumor cells abundantly release multiple classes of EVs. TMVs represent a discrete class of EVs separated from another well characterized class, exosomes, by their size (Figure 1a-c), cargo content (Supplementary Figure 1a), and distinct mode of biogenesis8,36. Furthermore, TMVs are distinct from apoptotic bodies, lacking cleaved forms of poly (ADP-ribose) polymerase-1 (PARP-1) or Lamin A/C, both of which become highly enriched during apoptotic cell death (Supplementary Figure 1b). Shed TMVs contain abundant bioactive cargos including numerous protein cargos (Figure 1d), and nucleic acids4, which led us to examine their endogenous miRNA content. Through bioanalyzer analysis, we identified small sized RNA cargo, similar in size to pre-miRNA, within TMVs shed from invasive melanoma (LOX), breast (MDA-MB-231), or prostate (PC-3) tumor cells (Figure 1e, Supplementary Figure 1c). Higher resolution analysis of also indicated the inclusion of mature miRNA-sized RNA cargo within melanoma TMVs (Figure 1f). Small RNAs, including miRNAs, were subsequently isolated, sequenced, and classified into small RNA pools including ribosomal RNA (rRNA), mitochondrial RNA (mtRNA), tRNA, and miRNA (Figure 1g). Inclusion of miRNAs as TMV cargo is of particular interest as miRNAs released from tumor cells impinge on various aspects of cancer pathogenesis and progression37-39.
Figure 1: TMVs are a distinct class of extracellular vesicles and contain pre-miRNA cargo.
Whole LOX melanoma cells (a), isolated LOX TMVs (b), or isolated LOX exosomes (c) were analyzed by scanning electron microscopy. Representative images of each population shown. Images are representative of N=3 biological and n=2 technical replicates. d. 20 μg of total protein isolate from whole cells (LOX Cell) or isolated TMVs (LOX TMV) was separated by SDS-PAGE and protein cargo determined by western blotting. Blots are representative of N=3 independent biological experiments. e. Agilent bioanalyzer analysis of TMV RNA content using the RNA 6000 Nano kit shows cargo corresponding in size to pre-miRNA (green bar). f. Higher resolution analysis of TMV small RNA using the Bioanalyzer Small RNA 6–150 nt Analysis kit shows peak corresponding to mature miRNA (green bar). Representative images from N=3 independent biological samples shown. g. Bowtie analysis of small RNA content isolated from TMVs released from invasive tumor cell lines of melanoma (LOX), prostate (PC-3), and breast (MDA-MB-231) origin. For each cell type, N=3 independent biological samples. h. ARF6 activity was measured using an MT2 ARF6-GTP specific pulldown assay as described in methods. Data presented is representative of N=4 independent biological experiments-. ARF6 i. Analysis of small RNA content isolated from parental melanoma cells or those expressing constitutively active ARF6-GTP showed no difference in the quantity of detectable RNA with ARF6 activation. j. Sequencing analysis revealed a significant increase in the total read hits corresponding to miRNA upon expression of ARF6-Q67L. k. qRT-PCR analysis confirms the increase in pre-miRNA cargo content in TMVs released by tumor cells of melanoma (LOX), ovarian (OvCar3), and breast (MDA-MB-231) origins. l. qRT-PCR analysis of total cell pre-miRNA levels with expression of ARF6-Q67L. Data presented as mean±standard deviation. For panels b-e, statistical analysis was based on measurements obtained for 3 biological repeats (N=3). P-values determined by unpaired, two-tailed t-test between control and treatment reactions for each independent experimental condition. P-values <0.05 were considered significant. Unprocessed blot images shown in Supplemental Image 7. Statistical Source in Supplementary Table 1.
ARF6 activation increases pre-miRNA and miRNA-related cargo in TMVs.
Given that ARF6 facilitates delivery of TMV cargo and also TMV release8, we examined whether activation of ARF6 led to changes in pre-miRNA and other miRNA related cargo contained within shed TMVs. Expression of constitutively active ARF6-Q67L led to a 5-fold increase in levels of active ARF6 (Figure 1h) and a 3-fold increase in TMV shedding (Supplementary Figure 2a). A comparison of total small RNA per TMV, however, showed no significant difference in ARF6-Q67L expressing cells relative to parental control cells, likely owing to the large proportion of non-miRNA content (Figure 1i). MicroRNA sequencing reads, however, showed a significant increase with ARF6 activation (Figure 1j) while a heat map of the 50 most abundant miRNAs found in TMVs revealed a global increase in TMV miRNA content upon ARF6 activation, highlighting that the observed increase was not due to enrichment of specific miRNAs (Supplementary Figure 2b). These results were confirmed by qRT-PCR of TMV pre-miRNA and mature miRNA targets selected for their pro-tumor roles, including oncomiR miR-2140; tumor-suppressor targeting miR-27a41 and miR-10042; and pro-migratory miR-15143 (Figure 1k, Supplementary Figure 2c). This increase in TMV miRNA content occurs despite a decrease in cellular levels of these miRNAs with activation of ARF6 (Figure 1l, Supplementary Figure 2d). Furthermore, as many biological effects of ARF6 are regulated through GDP/GTP cycling44, we confirmed these results using the fast-cycling mutant ARF6-T157N (Supplementary Figure 2e-g).
Exportin-5 is an ARF6 binding partner
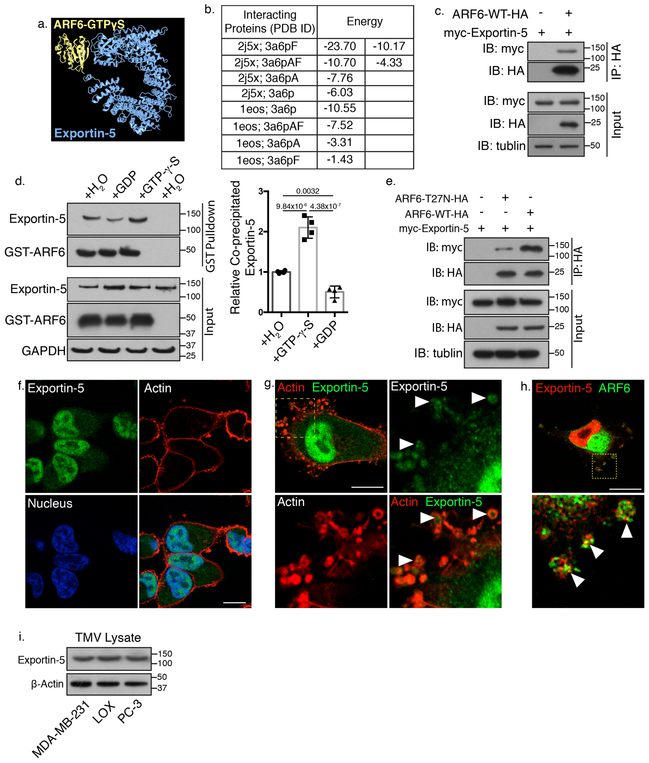
Increased TMV miRNA levels with ARF6 activation, suggested the active inclusion of miRNA within TMVs. Interrogation of publicly available databases for possible modes by which ARF6 could facilitate miRNA trafficking revealed that the pre-miRNA transporter, Exportin-5, a karyopherin known to transport pre-miRNA and RNA binding proteins across the nuclear envelope25,26,45, was an ARF6 binding partner identified through a large-scale mapping of protein-protein interactions by mass spectrometry46. Furthermore, the Protein Interactions by Structural Matching (PRISM) algorithm47 predicts a more favorable interaction between the Exportin-5/Ran-GTP/dsRNA (PDB ID 3a6p) complex and ARF6-GTP (2j5x), than with ARF6-GDP (1eos) (Figure 2a, b, Supplementary Figure 3a). This predicted interaction was validated through co-immunoprecipitation experiments (Figure 2c, Supplementary Figure 3b). A GTP-dependent interaction between Exportin-5 and ARF6 was confirmed via co-precipitation using recombinant ARF6 incubated with GTP-γ-S or GDP (Figure 2d), and through reciprocal co-immunoprecipitations from cells expressing dominant negative ARF6 (ARF6-T27N) which inhibits the interaction and reduces the amount of co-precipitating Exportin-5 (Figure 2e, Supplementary Figure 3c).
Figure 2: ARF6 interacts with the pre-miRNA transport protein Exportin-5.
a. PRISM prediction was used to model the predicted interaction between Exportin-5 and ARF6-GTPγS. b. Predicted binding energies between the Exportin-5 (3a6p) and ARF6-GTPγS (2j5x), or ARF6-GDP (1eos). c. Exportin-5 co-immunoprecipitates with ARF6 confirming the interaction predicted using PRISM. 200 μg of total protein was subjected to immunoprecipitation as described in methods. Antibody bound protein was precipitated using Protein-G conjugated Dynabeads before being resolved by SDS-PAGE and examined by western blotting. N=3 biologically independent experiments. d. Exportin-5 preferentially binds active, GTP-bound ARF6 in vitro. Recombinant GST-wt-ARF6 conjugated beads were incubated with lysates in the presence of 100 μM GTP-γ-S, 1 mM GDP, or vehicle control for 60 min at 37˚C. Bound proteins were precipitated and separated by SDS-PAGE for western blotting to examine relative amounts of co-precipitating Exportin-5. Data presented as mean±SD (N=4 biologically independent experiments). p-values determined by one-way ANOVA with Sidak’s correction for multiple comparisons. p-values <0.05 were considered significant. e. Dominant negative ARF6-T27N inhibits Exportin-5 co-precipitation. 200 μg of total protein from cells transiently expressing wt-ARF6 or ARF6-T27N was subjected to immunoprecipitation as described in methods. Immunoprecipitated protein was resolved by SDS-PAGE and examined by western blotting as indicated (N=3 biologically independent experiments). f. Immunofluorescent analysis of endogenous Exportin-5 in invasive melanoma cells reveals intracellular distribution divided between nuclear and cytoplasmic pools. Scale bar = 15 μm. g. Higher magnification analysis of Exportin-5 localization indicating the inclusion of Exportin-5 in nascent TMVs at the cell periphery (arrows). Scale bar = 15 μm. Panels f-g represent N=5 biologically independent experiments) h. Endogenous Exportin-5 and ARF6 co-localize in nascent TMVs at the cell periphery (arrows). Scale bar = 25 μm (N=3 biologically independent experiments) i. Western blot analysis of cargo content from TMVs isolated from multiple invasive cell lines confirms the inclusion of Exportin-5 as TMV cargo. (N=3 biologically independent experiments). Unprocessed blot images shown in Supplemental Image 7. Statistical Source in Supplementary Table 1.
Immunofluorescent analysis of endogenous Exportin-5 showed abundant Exportin-5 in the nucleus, together with a punctate cytoplasmic distribution consistent with chaperone protein cycling (Figure 2f). At higher magnification, Exportin-5 could be identified within budding TMVs (Figure 2g), where it co-localizes with ARF6 (Figure 2h, Supplementary Figure 3d). A portion of Exportin-5 also co-localizes with ARF6 in areas closely apposed to the nucleus (Supplementary Figure 3e). Additionally, Exportin-5 can be detected in isolated TMVs both by immunofluorescence (Supplementary Figure 3f) and western blotting (Figure 2i, Supplementary Figure 3g). Together these results suggest that, via association with Exportin-5, pre-miRNA cargo may continue to traffic from the nucleus to sites of TMV biogenesis for inclusion in shed TMVs.
MiRNA processing machinery is contained in shed TMVs
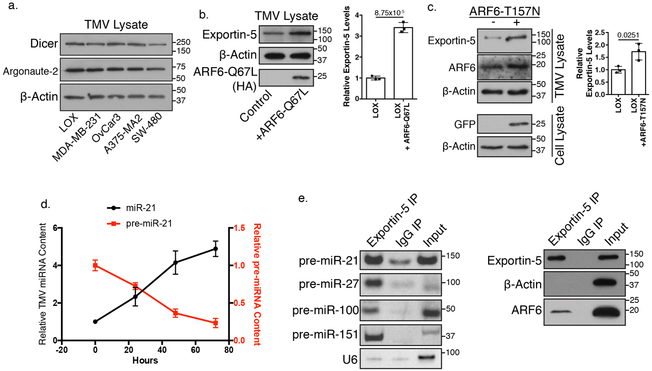
Exportin-5, Dicer, and Argonaute-2 were identified within shed TMVs (Figure 2i, 3a) and appeared enriched as a result of ARF6 activation (Figure 3b, c, Supplementary Figure 3h), indicating an important role for ARF6-regulated endosomal membrane trafficking in delivering miRNA related cargo to TMVs. Furthermore, the increase in mature TMV microRNA over time (Figure 3d), suggests that increases in mature miRNA with ARF6 activation stem from increased delivery of pre-miRNA. We then confirmed the inclusion of an Exportin-5/pre-miRNA/ARF6 complex in isolated TMVs (Figure 3e). While exosomes have been shown to contain pre-miRNA processing machinery14,35 its presence within TMVs was unknown. All of the above suggests that while there may be unique regulators in each EV pathway, the inclusion of RISC machinery within shed TMVs suggests this is likely a general phenomenon through which tumor cells modulate EV miRNA content.
Figure 3: pre-miRNA processing machinery is contained in shed TMVs.
a. TMVs from multiple invasive tumor cell lines were analyzed for the inclusion of pre-miRNA processing proteins Dicer and Argonaute-2 by western blotting. N=4 biologically independent samples. b. Western blot analysis of lysate generated from equal number of (1×108) TMVs released by parental melanoma cells compared to those expressing constitutively active ARF6 reveals an enrichment of Exportin-5 content within TMVs when ARF6 is activated. Data presented as mean±SD (N=3 biologically independent experiments). P-value determined by unpaired two-tailed t-test. P-value <0.05 was considered significant. c. Western blot analysis of lysate generated from equal number of (1×108) TMVs released by parental melanoma cells compared to those transfected with fast-cycling ARF6-T157N confirms an enrichment of Exportin-5 content within TMVs when ARF6 is activated. Data presented as mean±SD (N=3 biologically independent experiments). P-value determined by unpaired two-tailed t-test. p-value <0.05 was considered significant. d. RNA extracted from equal numbers (5×106) of isolated TMVs maintained in cell-free conditions at 37˚C for the times indicated was analyzed by qRT-PCR. The relative amounts of pre-miR21 and mature miR-21 were measured as described in methods. Data presented as mean±SD from N=3 biologically independent experiments. e. TMVs were isolated from invasive melanoma cells and endogenous Exportin-5 precipitated from 200 μg of input TMV protein. Co-precipitating RNA was examined by RT-PCR, and co-precipitating proteins examined by western blotting. Representative data from N=3 biologically independent samples shown. Unprocessed blot images shown in Supplemental Image 7. Statistical Source in Supplementary Table 1.
Casein kinase 2 facilitates cargo transfer from Ran to ARF6-GTP
Interphase cells maintain a strict Ran-GTP gradient across the nuclear envelope48. Following nuclear export, GTP hydrolysis on Ran is facilitated by the GTPase-activating protein, RanGAP1, which liberates cargo for further transport48. Given the previously reported activation of casein kinase 2 (CK2) downstream of ARF6-GTP49 and literature documenting that CK2-mediated 358S phosphorylation of RanGAP1 allows RanGAP1 to form a stable ternary complex with Ran-GTP and RanBP150, we examined the role of CK2 in Exportin-5 and pre-miRNA trafficking to shed TMVs. Treatment with the small molecule inhibitor of CK2 4,5,6,7-tetrabromobenzotriazole (TBB), reduces Exportin-5 included as shed TMV cargo (Figure 4a), along with a concomitant reduction in TMV pre-miRNA, and miRNA cargo (Figure 4b, Supplementary Figure 3i). TBB treatment did not change the levels of ARF6 incorporated into shed TMVs (Figure 4a) suggesting a specific disruption in cargo delivery rather than a global reduction in TMV cargo.
Figure 4: Casein Kinase 2 activity is needed for Exportin-5 trafficking.
a. Western blot analysis lysates generated from equal numbers of control or TBB treated cells (5×105) or TMVs (1×108). Data presented as mean±SD from N=3 biologically independent experiments. P-value determined by unpaired two-tailed t-test. P-value <0.05 was considered significant. b. RNA isolated from TMVs with or without TBB treatment was analyzed using qRT-PCR. CK2 inhibition results in a decrease in pre-miRNA contained within TMVs. Data presented as mean±SD for N=3 biologically independent experiments. P-values determined by unpaired, two-tailed t-test between control and treatment reactions for each independent pre-miRNA. P-values <0.05 were considered significant. c. Intracellular distribution of Exportin-5 was examined by immunofluorescence in cells treated with TBB or DMSO vehicle control. Scale bars = 15 μm. Representative images from N=3 biologically independent experiments shown. d. Heat map visualization of Exportin-5 channel described in c. e. LOX cells expressing constitutively active ARF6-Q67L were treated with TBB to block CK2 activity. The localization of Exportin-5 was then examined by immunofluorescent microscopy. Representative images (N=3 biologically independent experiments) shown. f. Heat map visualization of Exportin-5 channel described in e. g. Equal numbers of TMVs (1×108) were isolated from LOXARF6-GTP cells and subjected to western blot analysis to examine the levels of Exportin-5 contained as TMV cargo. Data presented as mean±SD (N=3 biologically independent experiments). p-value determined by unpaired two-tailed t-test. p-value <0.05 was considered significant. h. Melanoma cells expressing GFP-RanGAP were subjected to GFP-Trap and western blotting to examine changes in phosphoserine levels upon treatment with TBB. Representative data from N=3 biologically independent experiments shown. i. ARF6 was immunoprecipitated from cells with CK2 inhibition. The amount of Exportin-5 and Ran that co-precipitated with the GTPase was then examined by western blotting. Representative images from N=3 biologically independent experiments shown j. Exportin-5 was precipitated from cells treated with TBB and the levels of Ran and ARF6 which co-precipitated were subsequently examined by western blotting. Data is representative of N=3 biologically independent repeats. Unprocessed blot images shown in Supplemental Image 7. Statistical Source in Supplementary Table 1.
Intracellular distribution of Exportin-5 appeared perturbed upon TBB treatment, with CK2 inhibition leading to reduced cytoplasmic Exportin-5 and a concomitant increase in nuclear signal (Figure 4c, d). While we found no reduction in overall cellular levels of Exportin-5 (Figure 4a, Supplementary Figure 3j) a reduced cytoplasmic pool and increased nuclear pool were confirmed by cell fractionation (Supplementary Figure 3k). Similar results were obtained when CK2 was inhibited in the presence of constitutively active ARF6 (Figure 4e-g, Supplementary Figure 3l, m). With CK2 inhibition known to prevent 358S phosphorylation of RanGAP150, we examined RanGAP1 phosphoserine levels in the presence of TBB. Treatment with TBB reduces RanGAP p-serine levels when the GAP is isolated using a GFP trap (Figure 4h). Additionally, CK2 inhibition reduces the amount of both Ran and Exportin-5 that co-precipitate with ARF6 (Figure 4i) and leads to an increase in Ran and a decrease in ARF6 (Figure 4j) that co-precipitate with Exportin-5, suggesting that ARF6-mediated CK2 signaling facilitates the transfer of cargo from Ran to ARF6 for continued anterograde trafficking. Furthermore, examination of isolated TVM lysates revealed only small amounts of RanGAP and undetectable levels of Ran (Supplementary Figure 3n). Taken together these results reveal that CK2 activation downstream of ARF6-GTP is required to facilitate Exportin-5 and pre-miRNA trafficking to TMVs.
ARF6-mediated Exportin-5 and miRNA trafficking requires the cytohesin family of ARF-GEFs
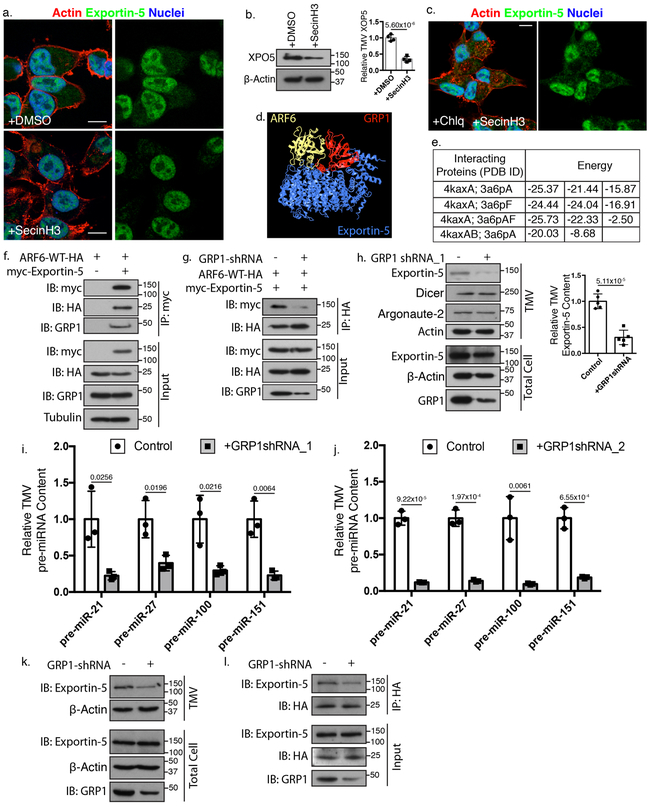
ARF6 is activated by guanine nucleotide exchange factors (GEFs)51 and inhibiting the cytohesin family of ARF GEFs using the small molecule inhibitor SecinH3 blocks ARF6-regulated functions in invasive tumor cells52. With SecinH3 treatment we observed a marked loss of Exportin-5 signal in the cytoplasm (Figure 5a) which was confirmed by western blotting of fractionated cells (Supplementary Figure 4a). Biochemical analysis confirmed the loss of Exportin-5 cargo in TMVs isolated from SecinH3-treated cells (Figure 5b). Of note, cytohesin inhibition did not result in a significant reduction of total Exportin-5 protein (Supplementary Figure 4b), or mRNA levels (Supplementary Figure 4c). Chloroquine treatment, was able to restore the cytoplasmic distribution of Exportin-5 (Figure 5c, Supplementary Figure 4d) suggesting that blockaded cargo, unable to be further trafficked is shunted to the lysosome for degradation. These results highlight the critical need for at least one functional member of the cytohesin family to facilitate anterograde Exportin-5 trafficking to sites of TMV biogenesis.
Figure 5: GRP1 scaffolding function facilitates Exportin-5 trafficking to TMVs.
a. Exportin-5 localization was examined by immunofluorescence in the presence of SecinH3. Scale bar = 15 μm. Representative images (N=5 biologically independent experiments shown. b. Exportin-5 western blot from equal numbers of TMVs from control or SecinH3 treated cells. Data presented as mean±SD for N=4 biologically independent experiments. P-value calculated using unpaired two-tailed t-test. c. Melanoma cells were co-treated with SecinH3 and chloroquine, and the intracellular distribution of Exportin-5 examined by confocal microscopy. Scale bar = 15 μm. Images representative of N=3 biologically independent experiments. d. Predicted interaction between Exportin-5 and ARF6 with GRP1 (4kax) was modeled, highlighting the energetically favorable binding arrangements listed in e. f. Exportin-5 co-immunoprecipitation from melanoma cells was analyzed by western blot to confirm the inclusion of GRP1 in complex with Exportin-5 and ARF6. Blots represent N=3 biologically independent experiments. g. Endogenous GRP1 was depleted from melanoma cells prior to immunoprecipitation of HA-ARF6. Co-precipitating proteins were separated by SDS-PAGE and analyzed by western blotting as indicated. Representative blots (N=3 biologically independent experiments) shown. h. Levels of Exportin-5, Dicer, and Argonaute-2 TMV cargo were analyzed by western blotting following isolation from cells depleted of endogenous GRP1. Data presented as mean±SD (N=5 biologically independent samples). P-value determined by unpaired two-tailed t-test. i, j. TMV pre-miRNA content was isolated from cells expressing 2 independent shRNA hairpins against GRP1. Isolated RNA was then analyzed by qRT-PCR. For each condition Data presented as mean±SD from 3 biologically independent experiments. P-values determined by unpaired, two-tailed t-test between control and treatment reactions for each independent pre-miRNA amplification reaction. k. Equal numbers of TMVs (1×108) were isolated from control or GRP1-shRNA treated LOXARF6-Q67L cells and lysates analyzed by western blotting as indicated. l. ARF6 was immunoprecipitated from LOXARF6-Q67L cells with or without GRP1-shRNA. Co-precipitating proteins were examined by western blot as indicated. For panels k-l representative blots (N=3 biologically independent experiments) shown. In all panels, p-values <0.05 were considered significant. Unprocessed blot images shown in Supplemental Image 7. Statistical Source in Supplementary Table 1.
To identify which cytohesin mediates Exportin-5 trafficking, we further interrogated publicly available databases and found that an ARF6-GTP and cytohesin 3 (GRP1) complex (PDB ID: 4kax) (Figure 5d), showed more favorable binding energies with Exportin-5 than the GTPase alone (Figure 5e). It is noteworthy that The Cancer Genome Atlas analysis53 of GRP1 expression revealed a strong negative correlation between higher than median expression levels and poor survival (Supplementary Figure 4e). Similar results were not seen with other cytohesin family members (Supplementary Figure 4f). A similar correlation between GRP1 expression and poor survival was found in specific tumor types including melanoma, ovarian cancer, breast cancer, squamous cell lung carcinoma, and bladder cancer (Supplementary Figure 5a-e), highlighting the importance of ARF activation during disease progression.
In light of the above, we identified an ARF6/Exportin-5/GRP1 complex through co-immunoprecipitation experiments (Figure 5f, Supplementary Figure 6a) which is disrupted by shRNA-mediated knockdown of GRP1 (Figure 5g, Supplementary Figure 6b). While depletion of GRP1 did not effect TMV shedding (Supplementary Figure 6c), it resulted in release of TMVs with decreased Exportin-5 though not Dicer, Argonaute-2 (Figure 5h, Supplementary Figure 6d), or other known TMV protein cargoes (Supplementary Figure 6e). Loss of GRP1 does reduce TMV pre-miRNA, and miRNA cargo (Figure 5i, j, Supplementary Figure 6f, g) despite increasing their cellular levels (Supplementary Figure 6h, i). Similar results were not seen with knockdown of cytohesins 1 or 2, the other cytohesins known to act on ARF654, where despite robust loss of the GEF, we found no change in levels of Exportin-5 TMV cargo (Supplementary Figure 6j).
The more favorable predicted binding energies, together with the known role for ARF6-GTP to bind GRP1 and relieve autoinhibition55, left open the possibility that in addition to acting as a GEF, GRP1 was also participating as a scaffold during Exportin-5 transfer from Ran-GTP to ARF6-GTP. Thus, we examined the impact of depleting GRP1 from cells with constitutive ARF6 activation. Loss of GRP1 led to a significant loss of Exportin-5 as TMV cargo (Figure 5k). Furthermore, GRP1 shRNA led to a reduction of Exportin-5 co-precipitating with ARF6 (Figure 5l). Thus, GRP1 scaffolding facilitates the interaction of ARF6-GTP and Exportin-5 and the interaction between ARF6 and the ARF-GEF GRP1 is critical for the anterograde trafficking of Exportin-5 and pre-miRNA cargo for inclusion in shed TMVs.
TMVs alter recipient cell behavior in part through the transfer of miRNA cargo
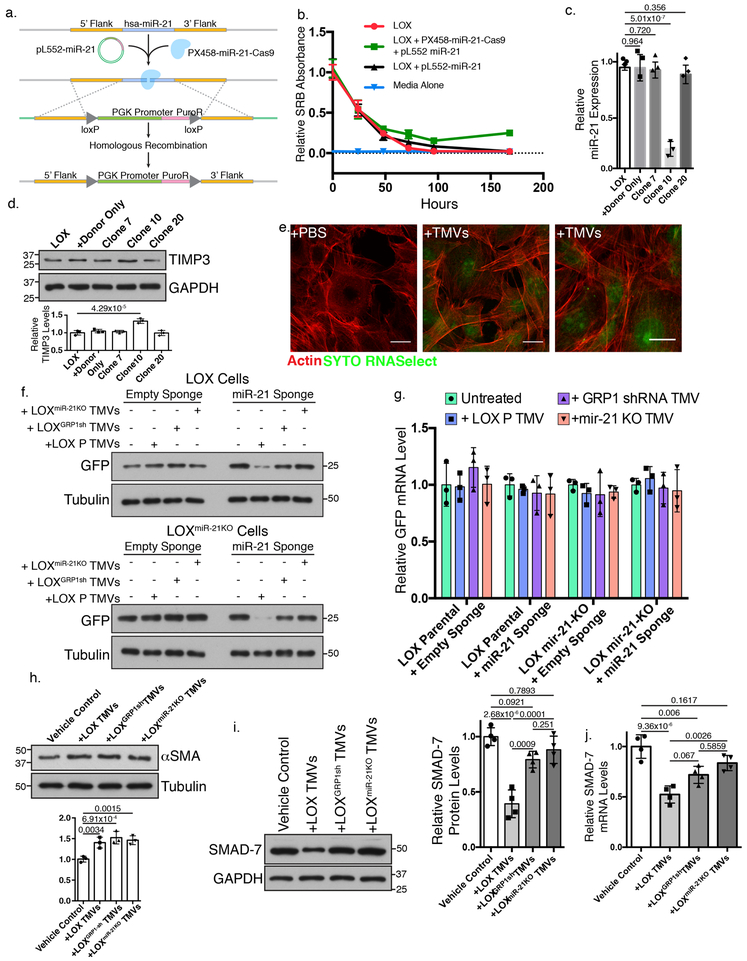
Shed TMVs, owing to their myriad bioactive cargoes, are capable of altering behavior in surrounding cells56. To examine the effect of TMV-mediated miRNA transfer, we focused on miR-21, and utilized a combination of CRISPR/Cas9 and homology directed repair (Figure 6a) to disrupt its genomic locus. Puromycin selection rapidly killed most cells before the candidate condition rebounded (Figure 6b). Examination of resulting cells revealed a population with significantly reduced miR-21 expression (Figure 6c) and corresponding increase in known miR-21 target TIMP-357 (Figure 6d). To examine the TMV-mediated RNA transfer we stained TMV contents with the cell-permeant RNA dye, SYTO RNAselect. Stained RNA is retained and distributed within recipient cells (Figure 6e). To monitor miRNA specific effects, we utilized miRNA sponge technology in which the 3’ UTR of destabilized GFP (dEGFP) contains miRNA binding sites to suppress translation58. TMV-transferred miR-21 effectively suppressed translation of dEGFP, leading to loss of protein at 10 hours. The loss of GFP protein was blocked by GRP1-shRNA or miR-21 depletion (Figure 6f). The loss of GFP protein occurred despite no loss of GFP mRNA levels at the early timepoint (Figure 6g). To examine TMV-mediated effects in an endogenous system, we incubated BJ human foreskin fibroblast cells with LOX melanoma TMVs. Incubation with TMVs lead to increased expression of alpha smooth muscle actin in recipient cells (Figure 6h). To examine miRNA-mediated effects, we examined the protein and mRNA levels of miR-21 target SMAD-759 and found that TMV-mediated miR-21 transfer leads to a loss of SMAD-7 protein and mRNA after 48 hours that is abrogated when shedding cells are depleted of GRP1 or miR-21 (Figure 6i, j). Taken together, these results highlight the functional consequences of TMV-mediated miRNA transfer downstream of GRP1 regulated trafficking.
Figure 6: TMVs transfer functional miRNAs to recipient cells.
a. Experimental scheme to knockdown endogenous miR-21 using HDR and CRISPR/Cas-9. b. 5,000 control; pL552-miR-21 transfected; or pL552-miR-21 and PX458-miR-21 dual transfected LOX cells were treated with 400 ng/mL puromycin (time t=0), and cell death measured. Data presented as mean±SD from 3 biologically independent samples. c. qRT-PCR of miR-21 in knockdown colonies. Data presented as mean±SD from 3 biologically independent samples. d. Western blotting of TIMP3 using 20 μg of lysate from control or miR-21 knockdown cells. Representative blots (N=3 biologically independent experiments) shown. e. Fibroblasts were incubated with SYTO RNAselect-stained TMVs for 24 hours before processing for confocal microscopy. Scale bar = 20 μm. Representative images (N=4 biologically independent experiments) shown. f. LOX or LOXmiR−21KO cells were transfected with miRNA sponges as indicated. Transfected cells were incubated with Isolated TMVs for 10 hours before being lysed and levels of dEGFP protein examined by western blot. Representative images shown (N=3 biologically independent experiments). g. LOX or LOXmiR−21KO cells were transfected with miRNA sponges and incubated with isolated TMVs for 10 hours before cells were lysed and dEGFP mRNA quantified by qRT-PCR. Mean±SD for N=3 biologically independent experiments shown. No statistically significant relationships were found. h. BJ fibroblasts were incubated with purified LOX TMVs for 48 hours and alpha smooth muscle actin levels measured by western blotting using 20 μg of cell lysates. Data presented as mean±SD (N=3 biologically independent samples). i. SMAD-7 levels were measured using 20 μg of BJ fibroblast cell lysate following treatment with isolated TMVs. Mean±SD (N=4 biologically independent experiments) shown. j. SMAD-7 mRNA levels in TMV-treated BJ fibroblasts were measured by qRT-PCR as described in methods. Data presented as mean±SD (N=4 biologically independent experiments). p-values determined by multiple unpaired two-tailed t-tests with Bonferroni’s correction (c, d, h); two-way ANOVA (g); or one-way ANOVA with Bonferroni’s correction (i, j).For all panels, p-values <0.05 were considered significant. Unprocessed blot images shown in Supplemental Image 7. Statistical Source in Supplementary Table 1.
Discussion and Conclusions:
The formation and release of tumor-derived microvesicles is a complex phenomenon with layered regulation governing TMV biogenesis and tightly controlling specific inclusion or exclusion of bioactive cargo. The findings presented here describe the events involved in the regulated trafficking of pre-miRNA cargo to sites of TMV biogenesis. This regulation occurs in part through formation of a trafficking complex containing Exportin-5, pre-miRNA, ARF6, and the ARF GEF, GRP1. In our working model for pre-miRNA trafficking, we propose that ARF6-GTP-regulated phosphorylation of RanGAP1 allows for the transfer of cargo exported from the nucleus into the ARF6-regulated shuttling complex for anterograde movement (Figure 8). Further, GRP1 serves as a scaffold facilitating the interaction between ARF6 and Exportin-5, supporting the contention that GEF’s have functional roles beyond nucleotide exchange60. While the mechanism outlined above results in the trafficking of the bulk of pre-miRNA cargo to TMVs, it does not exclude the possibility that further regulation may direct miRNA processing machinery; or specific miRNA or pre-miRNA sequences into TMVs, as has been proposed for exosome miRNA and mRNA cargo29,30,35. Indeed, the identification of CD43-regulated Dicer accumulation in cancer cell exosomes14 when combined with the results above showing differentual regulation of pre-miRNA and Dicer trafficking, certainly suggests this. These regulatory layers highlight the significant consequences stemming from TMV-mediated miRNA transfer as it provides a mechanism for direct alterations of gene expression, without further processing, allowing for rapid cellular responses in recipient cells.
Disassembly of Ran-GTP/Exportin-5 export complexes requires RanBP1 and RanGAP1 for GTP hydrolysis and cargo liberation. A similar interaction facilitated by RanBP2 in vertebrate cells releases cargo exported via interaction with Exportin-1 (Crm1) into the cytosol61. The strikingly similarity certainly elevates the possibility that these are biologically redundant alternatives utilized to export cargo from the nucleus to the cytosol. Our results, however, would suggest some level of specificity as we found no change in TMV Exportin-1 levels when inhibiting the pathway described above. This perhaps owing both to the soluble nature of RanGAP1 and RanBP1, and their high level of expression in tissue culture cells. Much Exportin-1 remains bound to the nuclear pore complex following transport complex disassembly and GTP hydrolysis, possibly for rapid shuttling back to the nucleus. In light of the results presented here it is tempting to speculate that it may also represent a level of cargo specificity with material needing to traffic longer distances being exported via Exportin-5. Alternatively, the pathway outlined above may represent an intermediate level of trafficking regulation falling between sequence specific targeting and the bulk movement of nuclear-associated cargo via the direct blebbing of the nuclear membrane62.
Research has suggested a global down-regulation of miRNA in tumors14,20,22. Of note, the majority of studies aimed at understanding this phenomenon have focused on disrupted miRNA biogenesis. It is conceivable that miRNA down regulation may be due to their expulsion, along with their precursors, as TMV cargo. Literature reports suggest that miRNAs contained within extracellular vesicles can influence gene expression in target cells63-65. The presence of mature miRNAs in TMVs underscores their propensity for manipulating normal tissues adjacent to the tumor producing tumor-induced “field effects”. Myriad biological processes in target cells are affected by EV miRNAs to, for example, prime metastatic sites66. The evidence presented outlining the regulated delivery of pre-miRNA cargo into structures with documented pro-tumorigenic roles, highlights the potential for the development of more tailored therapeutics by targeting such pathways.
Methods:
Antibodies and reagents:
Antibodies to myc tag, Exportin-5 (western blotting), β-actin, were purchased from Cell Signaling Technologies. Mouse monoclonal antibody to Exportin-5 used for immunofluorescence was obtained from Abnova. Antibodies against phospho-serine, Argonaute-2, alpha smooth muscle actin, and Dicer were purchased from Abcam. Antibody to GRP1(CYTH3) was purchased from GeneTex. Alpha-tubulin, MT1-MMP, and Fascin antibodies were purchased from Millipore Sigma. Mouse monoclonal anti-HA tag antibody was obtained from Covance. CYTH2,RhoA, Ran, and GAPDH antibodies were purchased from Santa Cruz Biotechnology. VAMP3 and Anti-GFP antibodies were purchased from ThermoFisher Scientific. Rab35, MHC-I, Rab22a, CYTH1, RanGAP, SMAD-7 and ARF6 (immunofluorescence) antibodies were purchased from Proteintech. TIMP3 antibody was purchased from R&D Technolgies. Mouse Monoclonal antibody to ARF6 was described previously67. Fluorophore conjugated secondary antibodies (donkey anti-mouse FITC, donkey anti-rabbit TRITC) were purchased from Jackson Immunoresearch. Antibody usage information is contained in Supplementary Table 2. SecinH3 and TBB were purchased from Cayman Chemical.
Short hairpin RNA:
shRNA against GRP1 (GRP1sh_1) was generated as previously published68. Briefly, a short hairpin RNA targeting GRP1 mRNA was subcloned into the lentiviral expression plasmid pLKO.1 using the following primer sequences: Fwd: 5’- CCGG—GCATTAAGAACGAGCCATTTA—CTCGAG—TAAATGGCTCGTTCTTAATGC—TTTTTG-3’; Rev: 5’- AATTCAAAAA— GCATTAAGAACGAGCCATTTA —CTCGAG— TAAATGGCTCGTTCTTAATGC −3’. The annealed oligos were further ligated between the AgeI and EcoRI sites of the pLKO.1 TRC-cloning vector. Positive clones were screened using EcoRI and NcoI double digestion and validated by sequencing. Additional MISSION shRNA constructs in the pLKO.1 background were purchased from Sigma-Aldrich. CYTH2:TRCN0000062100, CYTH1:TRCN0000062116, CYTH3:TRCN0000179183.
Cell Culture and transfection:
LOX melanoma cells (a gift from Prof. Oystein Fodstad, Oslo University) were cultured in RPMI (RPMI-1640 Life Technologies) supplemented with 10% (v/v) EquaFetal Serum (Atlas Biologicals), 2 mM L-glutamine and 100 U/mL penicillin-streptomycin (Life Technologies). LOXARF6-GTP cells were maintained similarly with the addition of 200 μg/mL Gentamycin and Hygromycin-B selection reagents. OvCar3 cells were maintained in RPMI supplemented with 20% EquaFetal Serum, 2 mM L-glutamine, 0.01 mg insulin, and 100 U/mL penicillin-streptomycin. PC-3 cells were grown in F-12K (Life Technologies) with 10% EquaFetal Serum and 100 U/mL penicillin-streptomycin. MDA-MB-231 and cells were grown in RPMI (Life Technologies) supplemented with 1 mM sodium pyruvate (Gibco), 10% EquaFetal Serum, 2 mM L-glutamine, and 100 U/mL penicillin-streptomycin. 293FT (Thermo Fisher Scientific) cells were maintained in culture according to the manufacturer’s specifications including supplementing with 0.5 mg/mL Geneticin (Thermo Fisher Scientific). All cell lines were maintained in humidified incubators with 5% CO2. In some experiments cells were pre-treated with .3 μM TBB (Cayman Chemical), 10 μM SecinH3 (Cayman Chemical), 50 μM chloroquine (Cell Signaling Technologies), or DMSO (Millipore Sigma) vehicle control for 6 hours before media replacement and continued treatment for an additional 24 hours. Where indicated, cells were transfected with indicated plasmids using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Lipofectamine:DNA ratios of 3:1 were utilized and scaled based on experimental needs, using 1 μg of plasmid DNA for 2×105 cells. To induce apoptosis, cells were treated with 50 nM okadaic acid (Millipore Sigma) for 24 hours.
Retro/lentiviral production and infection:
5 × 105 293FT cells were plated in 10 cm dishes in complete growth media without antibiotics and allowed to adhere for 24 hours at which time the plating media was replaced with 5 mL of complete media without antibiotics. Transfection mix was prepared using 9 μg of psPAX2, 3 μg of pMD2.G, 3 μg of lentiviral transfer plasmid, and 36 μL of Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Cells were incubated with transfection mix overnight and media changed within 16 hours. 10 mL of complete media without antibiotics was added and viral conditioned media harvested 48 hours post transfection. Supernatant was centrifuged at 3000 rpm for 15 minutes, filtered through 0.45 μm PVDF syringe filters, aliquoted, and stored at −80˚C until use. To infect tumor cells, viral aliquots were thawed on ice, mixed 1:1 with tumor growth media without antibiotics, and added to cells for 16 hours. Polybrene was added at a final concentration of 8 μg/μL at the time of infection.
TMV Isolation:
Cells for TMV isolation were plated to reach ~80% confluency at the time of conditioned media collection. Conditioned media was collected from cells, and the culture vessel rinsed 1x with sterile PBS which was subsequently added to the collected media. The mixture was then centrifuged for 15 minutes at 300 g to remove cells and large debris. The supernatant was then subjected to centrifugation for 20 minutes at 2,000 g to pellet apoptotic bodies. TMVs were then isolated by centrifugation at 10,000 g for 30 minutes. The resulting pellet was washed in sterile PBS 2 times before a final isolation prior to downstream use.
RNA isolation and cDNA generation:
TMVs, isolated, as described, were incubated with 2 mg/mL RNase A (Qiagen) for 30 minutes at 37˚C in PBS before being re-isolated for RNA extraction. Small RNA enriched total RNA was isolated from TMVs using the miRvana miRNA isolation kit (Thermo Fisher Scientific) according to manufacturer’s instructions. Isolated RNA was quantified using a NanoDrop 2000 (Thermo Fisher) to allow for input normalization in downstream applications. For mature miRNA analysis, 100 ng of isolated RNA was used as template for poly-A tailing reaction and cDNA generation using the qScript microRNA cDNA Synthesis kit (Quanta Bio). For pre-miRNA analysis, the cDNA synthesis method was adapted from previously published methods69. Briefly, a 15 μL reaction was assembled using 100 ng of isolated RNA, 300 nM pre-miRNA specific primers and primers for the RNU6B internal control. The mixure was then incubated at 80˚C for 5 minutes to denature the input RNA followed by 5 minutes at 60°C to anneal the primers and subsequently used for cDNA synthesis using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific) according to the manufacturer’s protocol. The pre-miRNA primer sequences were derived from previous research69. Pre-miR-21: F: 5’- GCTTATCAGACTGATGTTGACTG −3’; R: 5’- CAGCCCATCGACTGGTG −3’. Pre-miR-27a: F: 5’- GCAGGGCTTAGCTGCTTG −3’; R: 5’- GGCGGAACTTAGCCACTGT −3’. Pre-miR-100: F: 5’- AACCCGTAGATCCGAACTTG −3’; R: 5’- TACCTATAGATACAAGCTTGTGCG −3’. Pre-miR-151: F: 5’- CTCGAGGAGCTCACAGTCTAG −3’; R: 5’- GTCCTCAAGGAGCTTCAGTC −3’. RNU6B: F: 5’- CTCGCTTCGGCAGCACA −3’; R: 5’- AACGCTTCACGAATTTGCGT −3’.
TMV miRNA bioanalyzer analysis and sequencing:
Bioanalyzer small RNA analysis was carried out using the RNA 6000 Nano kit or the Small RNA Analysis kit on a 2100 Bioanalyzer Instrument (Agilent Technologies). For TMV miRNA sequencing, small RNA libraries were generated using the NEBNext Small RNA Library Prep Set for Illumina (New England BioLabs) followed by deep sequencing using the HiSeq 2500 (Illumina) platform. After the removal of adapter sequences, sequencing data were mapped to Homo sapiens miRNA sequence file collected from miRBase V21 for annotation using Bowtie2 and Samtools. Heat map comparison of the levels of 50 most abundant miRNAs in LOX TMVs and LOX-ARF6-GTP TMVs was performed using R library EdgeR after normalization by library size. R library gplots was then used to generate heatmap plot.
Quantitative Real-time PCR:
Quantitative RT-PCR (qRT-PCR) was performed on a Step-One-Plus real-time PCR machine (Applied Biosystems) and analyzed by the comparative CT method. Amplification was monitored using PerfeCTa SYBR Green SuperMix with ROX (Quanta Bio). Analysis of TMV miRNA levels was performed using miR specific primers purchased from Quanta Bio: hsa-miR-21–5p HSMIR-0021–5P; hsa-miR-100–5p HSMIR-0100–5P; hsa-miR-27a-3p HSMIR-0027A-3P; and hsa-miR-151a-3p HSMIR-0151A-3P. Pre-miRNA levels were analyzed using the same workflow, with the primers listed above. qRT-PCR to determine miR-21 knockdown was carried out as described above using Taqman probe (Hs04231424_s1) from ThermoFisher. EGFP (Mr04097229_mr) and SMAD-7 (Hs00998193_m1) levels were measured using the Fast Advanced Cells to CT kit from ThermoFisher. All Taqman assays used 18S ribosomal RNA (Hs99999901_s1) as input control and were cycled according to manufacturer’s instructions.
Immunofluorescence:
Cells were plated on glass coverslips, TMVs plated on poly-L-lysine coated coverslips, and allowed to adhere for a minimum of 18 hours before being fixed in 4% PFA (Electron Microscopy Supply). Fixed cells were washed 3 times with filtered 1X PBS; pre permeabilized with 1X PBS + 0.3% Triton X-100; blocked and permeabilized in 5% BSA, 0.2% Triton X-100, 0.05% Tween-20 in 1X PBS; and incubated with primary antibodies diluted in blocking buffer as indicated in figures. Cells were then incubated with fluorophore conjugated secondary antibodies together with rhodamine-phalloidin and ToPro-3 iodide (Life Technologies) and mounted using anti-fade gold mounting media (Life Technologies).
Scanning Electron Microscopy:
Cells, isolated TMVs or isolated exosomes were incubated overnight in naive EV-free culture media. For EV fractions, coverslips were first coated in poly-L-lysine (0.1% w/v). Samples were fixed in 2% electron microscopy grade glutaraldehyde (Electron Microscopy Sciences) diluted in SEM buffer (0.1M sodium cacodylate, pH 7.5) for 1 hour at room temperature. Samples were rinsed (3× 10 minutes) in SEM buffer before further fixation in 1% osmium tetroxide in SEM buffer for 1 hour at room temperature. Following a second rinsing step (3× 10 minutes) in fresh SEM buffer, samples were taken through a dehydration series of rinses in 50%, 70%, 80%, 95%, and 100% EM grade ethanol with each step consisting of 2 10-minute incubations. Samples in 100% ethanol were subjected to critical point drying by replacing ethanol with 100% liquid CO2 and bringing the CO2 to its critical point. Dried coverslips were mounted to SEM stubs (Ted Pella Inc.) using carbon tape and conductive silver paste. Samples were sputter coated with iridium to a thickness of 4 nm and imaged using a Magellan 400 (FEI) scanning electron microscope.
Immunoprecipitations and Western Blotting:
For western blotting, cells were lysed in buffer containing 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 20 mM Tris-HCl pH 7.5, and 1% Triton X-100 (all purchased from VWR). Just prior to use, mammalian protease inhibitor cocktail (Millipore Sigma) was added to complete the buffer. Triton insoluble fractions were removed by centrifugation at 15,000 g for 15 minutes. Lysates were then ready for use in downstream applications. For immunoprecipitation experiments, cells were first lysed in Co-IP buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 20 mM KCl, 1.5 mM MgCl2, and 0.5% Triton X-100. Just prior to use, mammalian protease inhibitor cocktail (Millipore Sigma) was added to complete the buffer. Insoluble fractions were removed by centrifugation as described above before being pre-cleared by incubation with Protein A/G Plus agarose beads (Santa Cruz Biotechnology) for 1 hour at 4˚C with gentle inversion. Pre-cleared lysates containing 200 μg total protein were subsequently transferred to new tubes for antibody incubation overnight at 4˚C with gentle inversion. Fresh Protein A/G Plus beads were added to the antibody mix and inverted for 1 hour at 4˚C. Beads were pelleted by centrifugation at 300 g for 2 minutes, supernatant removed, and 1 mL of fresh lysis buffer added. This was repeated for a total of 3 washes. Following the washing step, beads were resuspended in 1X SDS loading dye and incubated in a boiling water bath for 6 minutes before being separated by SDS-PAGE. PAGE gels were then transferred to PDVF membrane (Millipore Sigma). Non-specific binding was blocked by incubation in 5% nonfat milk in TBS + 0.05% Tween-20 for 1 hour at room temperature. Primary antibody incubation was carried out based on the manufacturer’s recommendations. HA immunoprecipitations were carried out using the Protein G Dynabead Immunoprecipitation Kit from ThermoFisher. Cells were lysed as described above before following the workflow outlined by the bead manufacturer.
ARF6-GTP Pulldown:
pGEX4T-MT2 plasmid was described previously67. Recombinant GST-MT2 was expressed in E-coli and conjugated to glutathione high capacity magnetic agarose beads (Millipore-Sigma) according to manufacturer’s instructions. LOX or LOX-ARF6-Q67L cells in log phase growth were lysed in MT2 assay buffer (50 mM Tris-HCl pH 7.4, 500 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, with mammalian protease inhibitor added just prior to use. Triton insoluble fractions were removed by centrifugation at 15,000 g for 15 minutes. For each pull-down assay, a 50-μl aliquot of the GST-MT2 fusion conjugated glutathione beads was washed three times with lysis buffer and incubated with 100 μg of lysate diluted to 200 μL in complete lysis buffer, with 2 mM ZnCl2 for, 1 hour at 4 °C with inversion. The beads were collected and washed three times with was buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 2 mM ZnCl2, and protease inhibitor. Bound proteins were eluted by boiling for 7 min in 1X SDS loading buffer and subjected to SDS-PAGE and western blotting.
In Vitro ARF6-Exportin-5 Pulldown:
pGEX-3X GST-wt-ARF6 plasmid was described previously. Recombinant ARF6 protein was conjugated to glutathione high capacity magnetic agarose beads according to manufacturer’s instructions. LOX cells, grown to 80% confluency, were lysed in co-immunoprecipitation buffer described above for 15 minutes at 4˚C and detergent insoluble fractions removed by centrifugation at 15,000 g for 15 minutes. While lysing cells, 50 μL of 50% bead slurry per sample were blocked in lysis buffer with 5% BSA for 10 minutes at 4˚C with gentle inversion. Following blocking, beads were rinsed twice with lysis buffer. 400 μg of total protein in pre-cleared lysates (diluted as needed to 1μg/μL with additional lysis buffer) were added to blocked beads together with 100 μM GTP-γ−S, 1 mM GDP, or vehicle control for 60 minutes at 37˚C. Following protein binding, beads were washed 3x with 1 mL of ice-cold lysis buffer, resuspended in 1X SDS sample buffer and boiled for 5 minutes, separated by SDS-PAGE, and probed by western blotting.
RNA Immunoprecipitation:
LOX cells were cultured in complete EV-free media for 72 hours prior to TMV isolation. TMV isolation was carried out as described above. Isolated TMVs were resuspended in sterile filtered PBS and incubated with 2 mg/mL RNase A (Qiagen) for 30 minutes at 37˚C in prior to being re-isolated and lysed in RIP buffer consisting of 150 mM KCl, 25 mM Tris pH 7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% Triton X-100 with protease inhibitor cocktail and 100 U/mL RNAase inhibitor SUPERase (Life Technologies) added immediately prior to use. The insoluble fraction was removed by centrifugation at 18,000 g for 15 minutes and the supernatant transferred to a new tube. Protein A/G Plus Agarose (Santa Cruz Biotechnology) bead slurry was added to the lysate (1:5 v/v) and the mixture incubated at 4˚C for 1 hour with inversion. Beads were removed and lysates were subsequently divided (Input, IgG IP, and Exportin-5 IP) and incubated with antibody overnight at 4˚C with gentle inversion before incubation with new Protein A/G Plus Agarose slurry for 1 hour. Beads were washed 3 times with fresh lysis buffer. After the 3rd wash, beads were resuspended in 1 mL fresh lysis buffer and 10% removed to validate IP by western blotting as described above. RIP beads were then washed once in sterile filtered PBS + SUPERase and protease inhibitor cocktail before being resuspended in cold Trizol. RNA was extracted following the manufacturer’s protocol. Precipitated RNA was then analyzed by PCR as described above. PCR products were separated on 5% non-denaturing TBE-PAGE gels, stained with ethidium bromide, and bands imaged.
CRISPR-Cas9-mediated Mir-21 Knockdown:
Guide RNA sequence targeting miR-21 (5’-CACCGCTCATGGCAACACCAGTCGA-3’) was selected from the GECKO library and cloned into the pSpCas9(BB)-2A-GFP (PX458) plasmid, a gift from Feng Zhang (Addgene plasmid # 48138; http://n2t.net/addgene:48138 ; RRID:Addgene_48138) as previously published70. Homology directed repair plasmid was based upon PL552, a gift from Su-Chun Zhang (Addgene plasmid # 68407; http://n2t.net/addgene:68407 ; RRID:Addgene_68407). Genomic DNA was isolated from exponentially growing LOX cells using the QIAamp DNA mini kit (Qiagen) and eluted into nuclease free water. 5’ and 3’ flanking regions of the miR-21 genomic locus were amplified by PCR using Phusion high-fidelity polymerase (Thermofisher) with the following primer pairs: miR-21-L-Kpn1–5.1 5’-GGCCAGTGAAGGTACCGTCTTACAAGTGAGCTGACACC-3’; miR-21-L-Sal1–3.1 5’-CACTGTAGAGGTACCGACATGGTGGAGTCGACTACTAC-3’; miR-21-R-BamH1–5.2 5’-CATTATACGGATCCATTTTGGTATCTTTCATCTGACCATC-3’; and miR-21-R-NotI-3.2 5’-TATCACCGGGCGGCCGCCAAGAAAGGAAGGCACGAACAG-3’; which were modified from previously published reports70. PCR products were cloned sequentially between the KpnI and SalI sites; and BamHI and NotI sites to generate pL552-miR-21. LOX cells were co-transfected with PX458 together with pL552-miR-21 using Lipofectamine-2000 (Thermofisher). Transfected cells were grown for 5 days before being selected with 400 ng/mL puromycin. Resulting colonies were screened for miR-21 levels using qRT-PCR.
Nanoparticle Tracking Analysis:
Conditioned media or TMVs were isolated from cells as described above. TMVs were resuspended in filtered PBS and injected into a NanoSight LM10 equipped with a 488 nm laser. Particles were tracked for 90 seconds, and tracking analysis carried out using NanoSight NTA software. Particles in suspension scatter a laser beam passing through the sample chamber in such a manner that they can easily be visualized at 20x magnification. The attached scientific CMOS camera captures a video file of the particles moving under Brownian motion. The software subsequently tracks many individual particles, estimating their hydrodynamic diameters using the Stokes-Einstein equation.
TMV-mediated MiRNA Transfer:
TMVs were isolated as described above, resuspended in filtered PBS, and quantified using NTA. Equal numbers of TMVs at a ratio of 50:1 (Figure 7f, g) or 25:1 (Figure 7e, h-j) to recipient cells, were pelleted and resuspended in naïve EV free cell culture media, and added to recipient cells in culture. Cells were incubated for 10 (Figure 7f, g) or 48 (Figure 7e, h-j) hours before use in downstream experimentation. 48-hour experiments were replenished with additional TMVs at t=24 hours. To stain endogenous RNA, TMVs or PBS vehicle control were stained in 500 nM SYTO RNAselect for 30 min at 37˚C. Stained TMVs were washed extensively in fresh filtered PBS before being resuspended in naïve EV-free cell culture media.
Statistics and Reproducibility:
Statistical analyses were performed using GraphPad Prism 6.0f or Microsoft Excel version 16.12. Student’s t test was used when comparing two groups with data that appeared to be normally distributed with similar variances. When comparing multiple treatment groups to a single control and the data were normally distributed, Bonferonni’s correction for multiple comparisons was employed. When multiple groups were being analyzed and each group was compared to all other groups and the data were normally distributed, we performed a one-way ANOVA with Sidak’s multiple comparisons correction. No outliers were exclude from analysis and all experiments were performed on at least 3 biological replicates under similar conditions.
Data Availability:
Deep-sequencing (miRNA–seq) data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE130316. Previously published protein-protein interaction data that was analyzed for this manuscript is available through Intact database (www.ebi.ac.uk/intact), accession EBI-105937047, pubid:1735393171 (Figures 2b, and 5e). Data used for PRISM predictions was accessed through (http://cosbi.ku.edu.tr/prism/) using structures stored in the RCSB Protein Data Bank (https://www.rcsb.org/) (Figures 2a, 5d, and S3a). The Cancer Genome Atlas data analyzed in this manuscript was accessed via the Xena Functional Genomics Explorer72 (www.xenabrowser.net) (Figures S4e and S4f; and S5). All other data generated for and analyzed in this manuscript is available by contacting the corresponding author. Source data for Figures 1-6 and Supplementary Figures 2-4, and 6, have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements:
This work was supported in part by grants from the National Cancer Institute (R01CA115316) and the Catherine Peachey Foundation to C.D-S.
Footnotes
Financial Statement. The authors have no financial or non-financial competing interests.
References:
- 1.Abels ER & Breakefield XO Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol Neurobiol 36, 301–312, doi: 10.1007/s10571-016-0366-z (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desrochers LM, Antonyak MA & Cerione RA Extracellular Vesicles: Satellites of Information Transfer in Cancer and Stem Cell Biology. Dev Cell 37, 301–309, doi: 10.1016/j.devcel.2016.04.019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tricarico C, Clancy J & D’Souza-Schorey C Biology and biogenesis of shed microvesicles. Small GTPases, 1–13, doi: 10.1080/21541248.2016.1215283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Niel G, D’Angelo G & Raposo G Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19, 213–228, doi: 10.1038/nrm.2017.125 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Clancy JW et al. Regulated delivery of molecular cargo to invasive tumour-derived microvesicles. Nat Commun 6, 6919, doi: 10.1038/ncomms7919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sedgwick AE, Clancy JW, Olivia Balmert M & D’Souza-Schorey C Extracellular microvesicles and invadopodia mediate non-overlapping modes of tumor cell invasion. Sci Rep 5, 14748, doi: 10.1038/srep14748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazaro-Ibanez E et al. Metastatic state of parent cells influences the uptake and functionality of prostate cancer cell-derived extracellular vesicles. J Extracell Vesicles 6, 1354645, doi: 10.1080/20013078.2017.1354645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muralidharan-Chari V et al. ARF6-Regulated Shedding of Tumor Cell-Derived Plasma Membrane Microvesicles. Current Biology 19, 1875–1885, doi: 10.1016/j.cub.2009.09.059 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Souza-Schorey C & Clancy JW Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev 26, 1287–1299, doi: 10.1101/gad.192351.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T et al. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc Natl Acad Sci U S A 111, E3234–3242, doi: 10.1073/pnas.1410041111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabhan JF, Hu R, Oh RS, Cohen SN & Lu Q Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A 109, 4146–4151, doi: 10.1073/pnas.1200448109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q & Lu Q Plasma membrane-derived extracellular microvesicles mediate non-canonical intercellular NOTCH signaling. Nat Commun 8, 709, doi: 10.1038/s41467-017-00767-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo L & Freed EO ARRDC1 as a mediator of microvesicle budding. Proc Natl Acad Sci U S A 109, 4025–4026, doi: 10.1073/pnas.1201441109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melo SA et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721, doi: 10.1016/j.ccell.2014.09.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menck K et al. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J Mol Cell Biol 7, 143–153, doi: 10.1093/jmcb/mju047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skog J et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology 10, 1470–1476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sansone P et al. Evolution of Cancer Stem-like Cells in Endocrine-Resistant Metastatic Breast Cancers Is Mediated by Stromal Microvesicles. Cancer Res 77, 1927–1941, doi: 10.1158/0008-5472.CAN-16-2129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC & Rak J Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A 106, 3794–3799, doi:0804543106 [pii] 10.1073/pnas.0804543106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Nedawi K et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature Cell Biology 10, 619–624 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Calin GA & Croce CM MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–866, doi: 10.1038/nrc1997 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Giovannetti E et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res 70, 4528–4538, doi: 10.1158/0008-5472.CAN-09-4467 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Melo SA & Esteller M Disruption of microRNA nuclear transport in human cancer. Semin Cancer Biol 27, 46–51, doi: 10.1016/j.semcancer.2014.02.012 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Jiang Q et al. MicroRNA-100 suppresses the migration and invasion of breast cancer cells by targeting FZD-8 and inhibiting Wnt/beta-catenin signaling pathway. Tumour Biol 37, 5001–5011, doi: 10.1007/s13277-015-4342-x (2016). [DOI] [PubMed] [Google Scholar]
- 24.Song B et al. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res 29, 29, doi: 10.1186/1756-9966-29-29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohnsack MT, Czaplinski K & Gorlich D Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10, 185–191 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi R, Qin Y, Macara IG & Cullen BR Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17, 3011–3016, doi: 10.1101/gad.1158803 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slezak-Prochazka I, Durmus S, Kroesen BJ & van den Berg A MicroRNAs, macrocontrol: regulation of miRNA processing. RNA 16, 1087–1095, doi: 10.1261/rna.1804410 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei Z et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun 8, 1145, doi: 10.1038/s41467-017-01196-x (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S & Schekman R Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5, doi: 10.7554/eLife.19276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villarroya-Beltri C et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4, 2980, doi: 10.1038/ncomms3980 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosaka N et al. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem 288, 10849–10859, doi: 10.1074/jbc.M112.446831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosaka N et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 285, 17442–17452, doi: 10.1074/jbc.M110.107821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guduric-Fuchs J et al. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics 13, 357, doi: 10.1186/1471-2164-13-357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibbings DJ, Ciaudo C, Erhardt M & Voinnet O Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11, 1143–1149, doi: 10.1038/ncb1929 (2009). [DOI] [PubMed] [Google Scholar]
- 35.McKenzie AJ et al. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep 15, 978–987, doi: 10.1016/j.celrep.2016.03.085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D’Souza-Schorey C & Clancy JW Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes & development 26, 1287–1299, doi: 10.1101/gad.192351.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iorio MV & Croce CM MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4, 143–159, doi: 10.1002/emmm.201100209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croce CM Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10, 704–714, doi: 10.1038/nrg2634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin S & Gregory RI MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15, 321–333, doi: 10.1038/nrc3932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medina PP, Nolde M & Slack FJ OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467, 86–90, doi: 10.1038/nature09284 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Guttilla IK & White BA Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem 284, 23204–23216, doi: 10.1074/jbc.M109.031427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng YS et al. MiR-100 regulates cell differentiation and survival by targeting RBSP3, a phosphatase-like tumor suppressor in acute myeloid leukemia. Oncogene 31, 80–92, doi: 10.1038/onc.2011.208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding J et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol 12, 390–399, doi: 10.1038/ncb2039 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Klein S, Franco M, Chardin P & Luton F Role of the Arf6 GDP/GTP cycle and Arf6 GTPase-activating proteins in actin remodeling and intracellular transport. J Biol Chem 281, 12352–12361, doi: 10.1074/jbc.M601021200 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Brownawell AM & Macara IG Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol 156, 53–64, doi: 10.1083/jcb.200110082 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ewing RM et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3, 89, doi: 10.1038/msb4100134 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keskin O, Nussinov R & Gursoy A PRISM: protein-protein interaction prediction by structural matching. Methods Mol Biol 484, 505–521, doi: 10.1007/978-1-59745-398-1_30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart M Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol 8, 195–208, doi: 10.1038/nrm2114 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Pellon-Cardenas O, Clancy J, Uwimpuhwe H & D’Souza-Schorey C ARF6-regulated endocytosis of growth factor receptors links cadherin-based adhesion to canonical Wnt signaling in epithelia. Mol Cell Biol 33, 2963–2975, doi: 10.1128/MCB.01698-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda E, Hieda M, Katahira J & Yoneda Y Phosphorylation of RanGAP1 stabilizes its interaction with Ran and RanBP1. Cell Struct Funct 30, 69–80 (2005). [DOI] [PubMed] [Google Scholar]
- 51.D’Souza-Schorey C & Chavrier P ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 7, 347–358 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Grossmann AH et al. The small GTPase ARF6 stimulates beta-catenin transcriptional activity during WNT5A-mediated melanoma invasion and metastasis. Sci Signal 6, ra14, doi: 10.1126/scisignal.2003398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldman M et al. The UCSC Xena Platform for cancer genomics data visualization and interpretation. bioRxiv, doi: 10.1101/326470 (2018). [DOI] [Google Scholar]
- 54.Hongu T & Kanaho Y Activation machinery of the small GTPase Arf6. Adv Biol Regul 54, 59–66, doi: 10.1016/j.jbior.2013.09.014 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Donaldson JG & Jackson CL ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 12, 362–375, doi: 10.1038/nrm3117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clancy JW, Tricarico CJ, Marous DR & D’Souza-Schorey C Coordinated Regulation of Intracellular Fascin Distribution Governs Tumor Microvesicle Release and Invasive Cell Capacity. Mol Cell Biol 39, doi: 10.1128/MCB.00264-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin del Campo SE et al. MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: in vivo effects of MiR-21 inhibitor. PLoS One 10, e0115919, doi: 10.1371/journal.pone.0115919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ebert MS, Neilson JR & Sharp PA MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4, 721–726, doi: 10.1038/nmeth1079 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Q et al. MiR-21/Smad 7 signaling determines TGF-beta1-induced CAF formation. Sci Rep 3, 2038, doi: 10.1038/srep02038 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cohen LA et al. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell 18, 2244–2253, doi: 10.1091/mbc.e06-11-0998 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritterhoff T et al. The RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3 ligase is a disassembly machine for Crm1-dependent nuclear export complexes. Nat Commun 7, 11482, doi: 10.1038/ncomms11482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reis-Sobreiro M et al. Emerin Deregulation Links Nuclear Shape Instability to Metastatic Potential. Cancer Res 78, 6086–6097, doi: 10.1158/0008-5472.CAN-18-0608 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Thind A & Wilson C Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles 5, 31292, doi: 10.3402/jev.v5.31292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dror S et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat Cell Biol 18, 1006–1017, doi: 10.1038/ncb3399 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Sakha S, Muramatsu T, Ueda K & Inazawa J Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep 6, 38750, doi: 10.1038/srep38750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104, doi: 10.1038/nature15376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schweitzer JK & D’Souza-Schorey C Localization and activation of the ARF6 GTPase during cleavage furrow ingression and cytokinesis. J Biol Chem 277, 27210–27216, doi: 10.1074/jbc.M201569200 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Hongu T et al. Arf6 regulates tumour angiogenesis and growth through HGF-induced endothelial beta1 integrin recycling. Nat Commun 6, 7925, doi: 10.1038/ncomms8925 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Jiang J, Lee EJ, Gusev Y & Schmittgen TD Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res 33, 5394–5403, doi: 10.1093/nar/gki863 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87, doi: 10.1126/science.1247005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ewing RM et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3, 89, doi: 10.1038/msb4100134 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goldman M et al. The UCSC Xena Platform for cancer genomics data visualization and interpretation. bioRxiv, doi: 10.1101/326470 (2018). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep-sequencing (miRNA–seq) data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE130316. Previously published protein-protein interaction data that was analyzed for this manuscript is available through Intact database (www.ebi.ac.uk/intact), accession EBI-105937047, pubid:1735393171 (Figures 2b, and 5e). Data used for PRISM predictions was accessed through (http://cosbi.ku.edu.tr/prism/) using structures stored in the RCSB Protein Data Bank (https://www.rcsb.org/) (Figures 2a, 5d, and S3a). The Cancer Genome Atlas data analyzed in this manuscript was accessed via the Xena Functional Genomics Explorer72 (www.xenabrowser.net) (Figures S4e and S4f; and S5). All other data generated for and analyzed in this manuscript is available by contacting the corresponding author. Source data for Figures 1-6 and Supplementary Figures 2-4, and 6, have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.