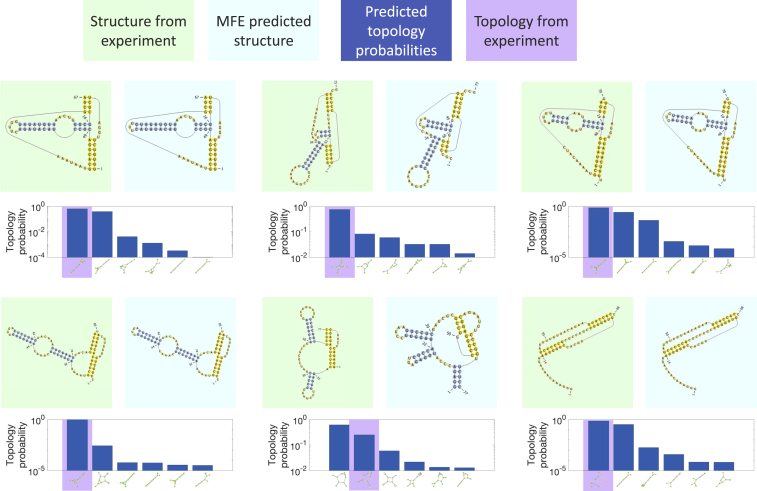
Figure 6.
Comparison to experiments for long sequences. Six long sequences were chosen from the PseudoBase++ database as described in the main text. The sequences are fragments derived from the following (starting from the top left and moving across): tobacco mosaic virus (124, 125, 126), Bacillus subtilis (127), tobacco mild green mosaic virus (125, 128), Bacillus subtilis (129), Giardiavirus (130), and Visna-Maedi virus (131). The experimental structures are supported by (numbering the sequences in the same order) sequence comparison (1–4,6), structure probing (1,3,5,6), mutagenesis (2,4–6), three-dimensional modeling (1), and NMR (6). We show the experimental structure (green background) and the MFE-predicted structure (light blue background) plotted using the PseudoViewer software (90). We also display the top six topologies (out of several hundred, depending on the particular sequence) and their respective predicted probabilities, with the topology corresponding to the experimental structure highlighted in purple. Overall, our results demonstrate successful predictions even for these long pseudoknotted sequences, especially in terms of the predicted topology. To see this figure in color, go online.