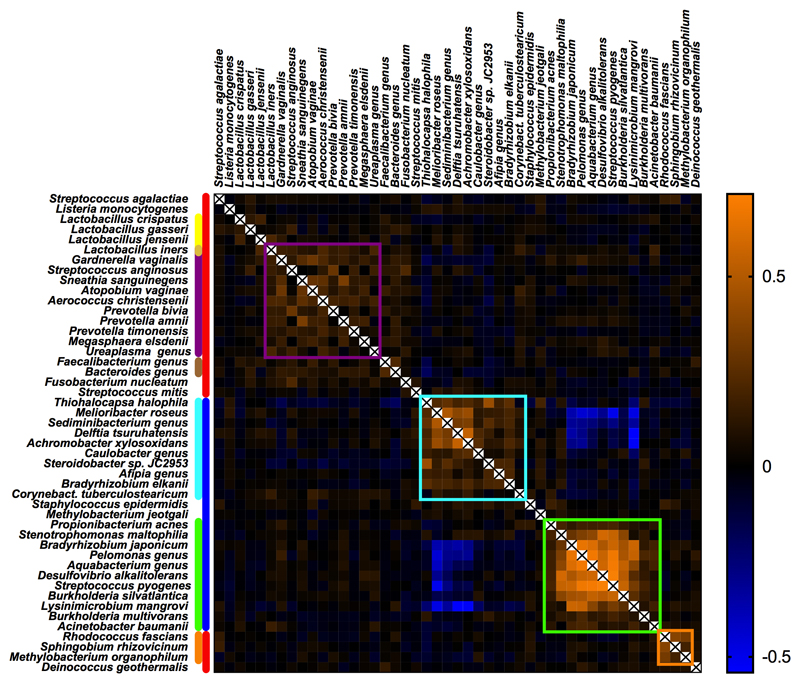
Extended Data Fig. 8. Heatmap of Spearman’s rho correlation coefficients of bacterial signals as found by 16S rRNA amplicon sequencing.
Sample-associated signals (red bar), are typically identified by elevated kappa scores as shown in Supplementary Table 4. Reagent contaminants are indicated with a blue bar. Vaginosis associated bacteria (purple bar) show positive correlations (purple square) with each other, Lactobacillus iners and fecal bacteria (brown bar). Lactobacilli (yellow bar) show limited positive correlation with fecal bacteria. Reagent contaminants mainly associated with the Qiagen (light blue) or the Mpbio kit (green) form distinct clusters. Species which are strongly associated with sample collection contamination in 2012-2013 are indicated in orange. For each species the highest value (%) found using either the Qiagen or the Mpbio DNA isolation kit, was used as input (using all 498 samples).