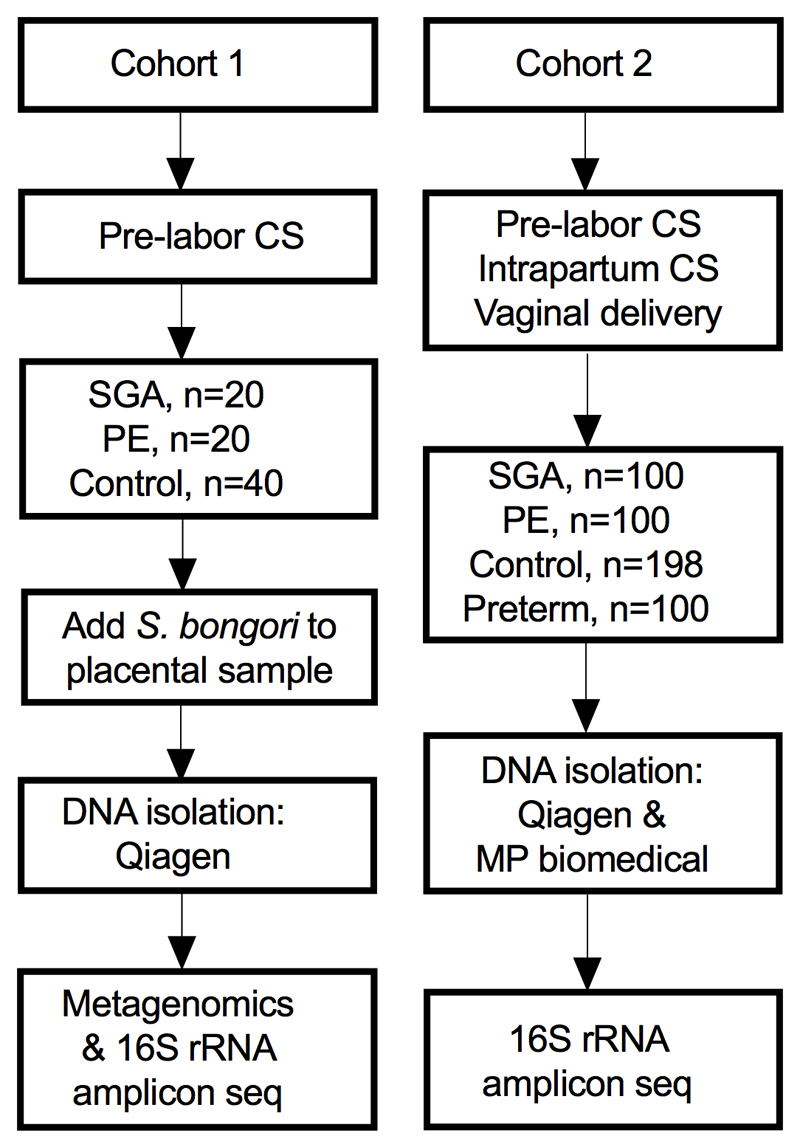
Extended Data Fig. 1. Two cohorts of placental samples were analysed.
Cohort 1 (n=80) contained only samples from pre-labour CS and S. bongori was added to the samples before DNA isolation as a positive control. The samples in cohort 1 was analysed by both metagenomics as well as by 16S rRNA amplicon sequencing. Cohort 2 (n=498) contained placental samples from CS and vaginal deliveries. DNA was isolated twice from each placental sample with two different DNA extraction kits. The samples were analysed by 16S rRNA amplicon sequencing. CS = Caesarean section, SGA = small for gestational age (birth weight <5th percentile) using a customized reference, PE = preeclampsia using the ACOG 2013 definition, Preterm= birth at <37 week’s gestation.