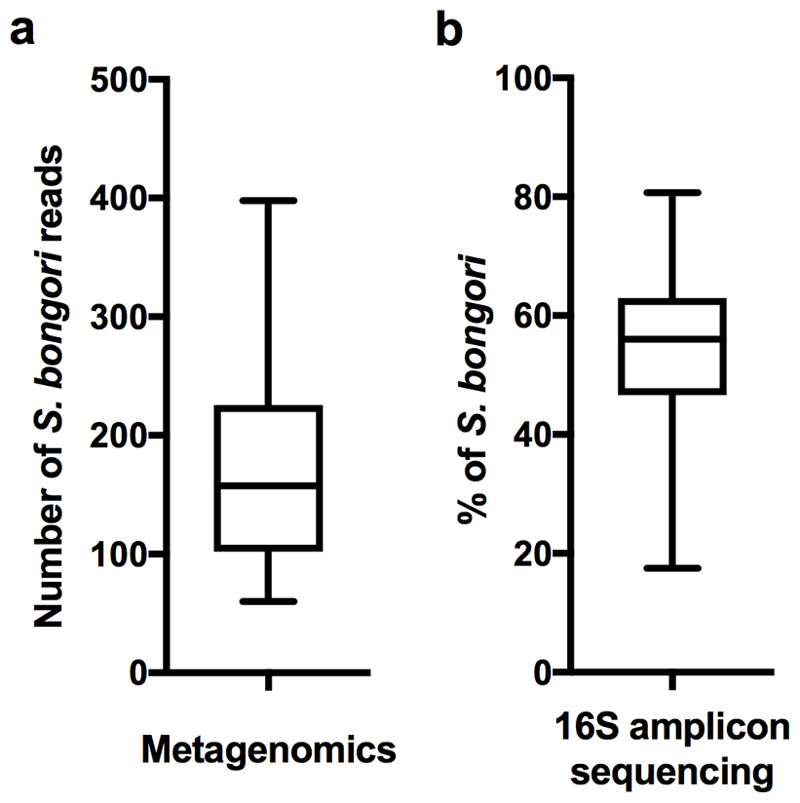
Extended Data Fig. 2. Positive control experiment comparison between metagenomics and 16S amplicon sequencing.
Adding approximately 1,100 CFU of S. bongori to the placental tissue before DNA isolation resulted in a) an average of 180 reads (SD: 90 reads) by metagenomic sequencing (n=80) or b) on average 54% of all 16S rRNA amplicon sequencing reads (~33,000 reads) being identified as S. bongori (SD: 13%; n=79). Box represents the interquartile range. Whiskers represent Max/Min in both figures.