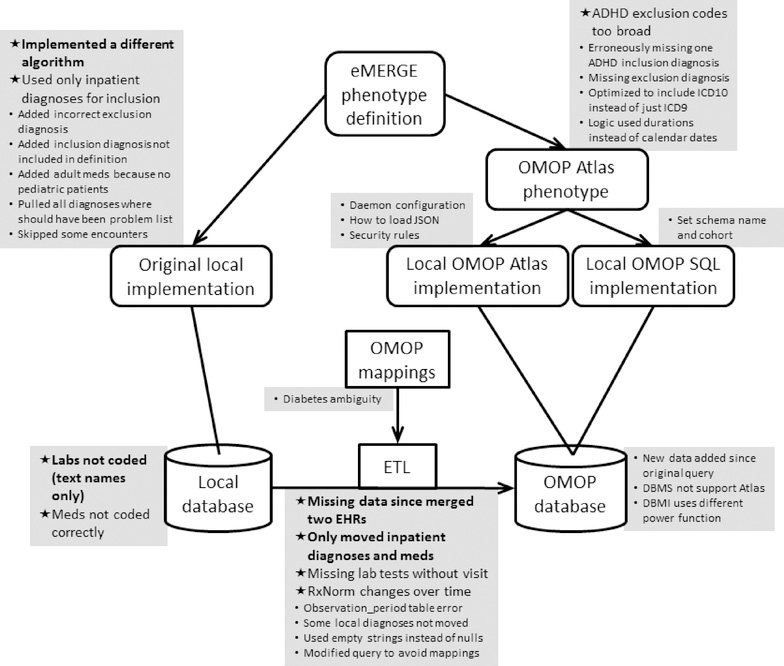
Figure 1. Phenotyping flowchart and issues reported.
Phenotypes were based on the published eMERGE phenotype definition, which included a narrative definition, high-level concept code lists, a flowchart, and pseudocode, and it was taken as the gold standard definition and therefore had no issues. The original eMERGE implementation had each local site write software to query their local database based on the eMERGE definition. In this study, each site converted its data to the OHDSI OMOP database using extract-transfer-load (ETL) software and OMOP vocabulary mappings. The eMERGE definition was encoded as a single OMOP phenotype using the OHDSI Atlas tool. A site could use a local copy of Atlas running on their OMOP database to run the phenotype or use SQL that was generated automatically by Atlas for five database management systems. Where possible, the OMOP result of the phenotype query was compared to the original result. Sites reported issues that they encountered, which are shown in grey squares adjacent to the most relevant step. Issues that caused a significant difference between the original and OMOP query are marked with a star in bold. Those that caused a moderate difference are marked with a star in non-bold. Issues that caused little or no change are marked with a smaller round bullet in a smaller font.