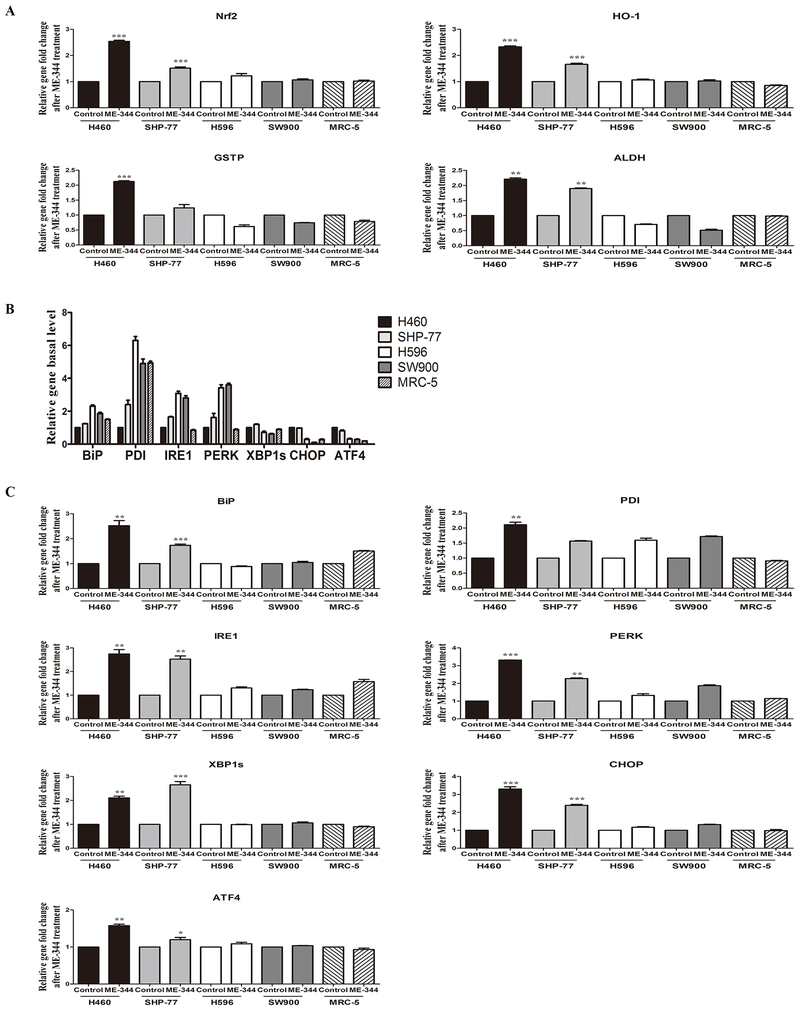
Figure 3. ME-344-induced changes in gene expression of Nrf2, HO-1, GSTP, ALDH and UPR markers in sensitive (H460 and SHP-77) and resistant (H596 and SW900) human lung cancer and normal cells (MRC-5).
Cells were treated with their IC50s for 24 h. A. Fold-changes in gene expression (Nrf2, HO-1, GSTP, ALDH) after ME-344 treatment relative to untreated control cells with mean values set at 1. Relative gene expression quantification was based on the comparative threshold cycle (CT) method (2-ΔΔCt) with normalization of the raw data to the included housekeeping gene (18S rRNA). B. Basal gene expression (UPR markers) was analyzed by real-time PCR in untreated control cells. Relative gene expression quantification was based on the comparative threshold cycle (CT) method (2-ΔΔCt) with normalization of the raw data to the included housekeeping gene (18S rRNA) and relative to H460 cell line with mean values set at 1. C. Fold-changes in gene expression (UPR markers) after ME-344 treatment relative to untreated control cells with mean values set at 1. Relative gene expression quantification was based on the comparative threshold cycle (CT) method (2-ΔΔCt) with normalization of the raw data to the included housekeeping gene (18S rRNA). Data are derived from three independent experiments and presented as means ± SEM in the bar graphs. *p < 0.05, **p < 0.01, ***p < 0.001 vs. the untreated control by Student’s t-test.