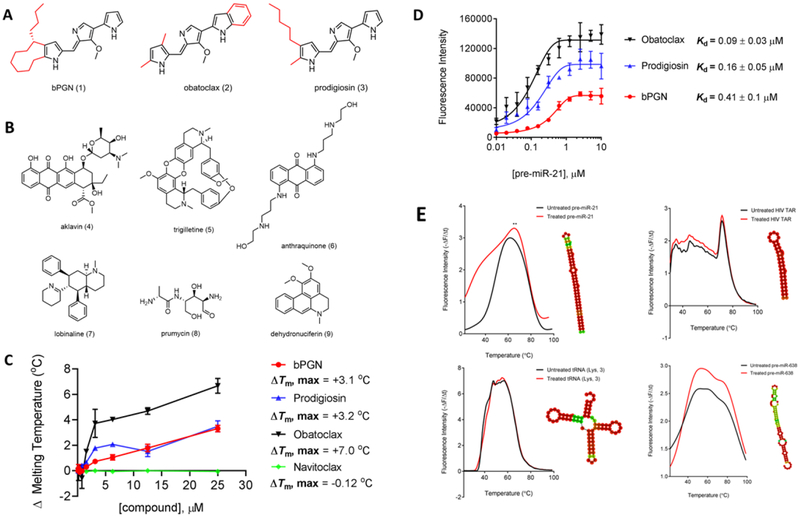
Figure 1: Identification of RNA-binding compound, bPGN (1).
(A) The chemical structures of prodiginine-class compounds: butylcycloheptyl prodiginine (1), obatoclax (2), and prodigiosin (3). (B) The chemical structures of active natural product compounds identified through a DSF-based HTS against pre-miR-21. (C) Titration of 1, 2, 3, and navitoclax ranging from 0 – 25 μM to pre-miR-21 using DSF shows melting temperature (Tm) changes. (D) Direct binding affinity of 1, 2, 3, was determined by titrating compounds from 0 – 10 μM with pre-miR-21. Fluorescence was monitored at 545/580 nm. Kd was assessed using the non-linear regression fit (Sigmoidal, 4PL) function in GraphPad Prism. (E) Pre-miR-21, tRNA (Lys, 3), HIV TAR, and pre-miR-638 were incubated with 5 μM of 1 for 10 minutes followed by a Tm shift measurement by DSF. Only pre-miR-21 showed significant Tm shift. Interestingly, pre-miR-628 showed significant fluorescence intensity change, but no Tm shift was observed. All experiments were performed in triplicate.