Fig. 2.
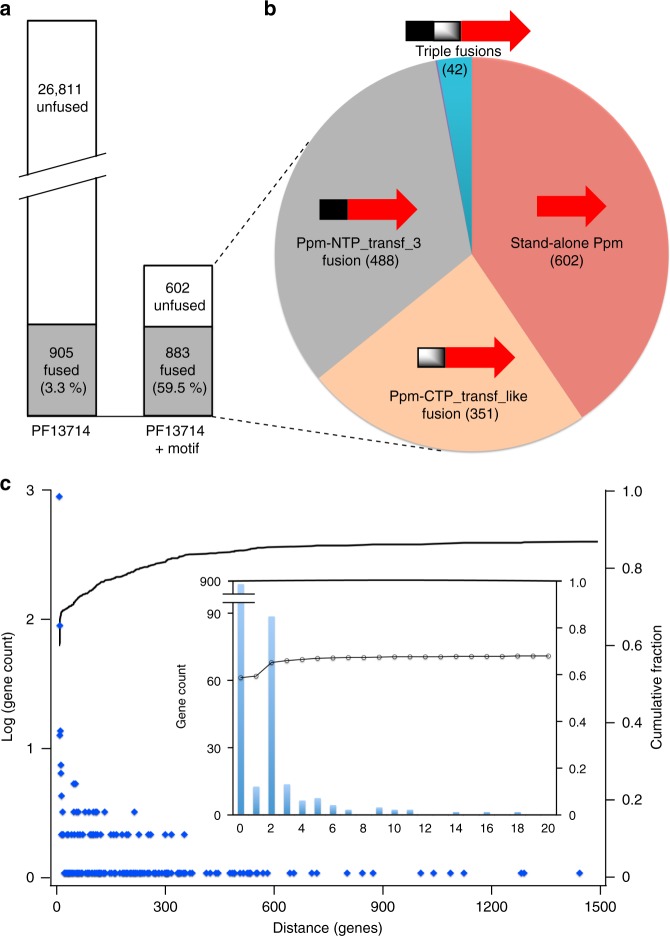
Genomic relationships between ppm and nucleotidyltransferase-encoding genes. a Left: 27,716 non-redundant Ppm protein sequences were identified in NCBI’s refSeq prokaryotic genome database that matched the PF13714 HMM, of which 905 were fused to other domains. Right: After filtering for the presence of the characteristic Ppm motif (EDKXXXXXNS), 26,231 presumed non-Ppm sequences were removed. Only 22 of these removed sequences were fused to other domains. b Summary of the remaining 1485 predicted Ppm proteins and inventory of fusion status: Ppm alone (red, 40.5%); PF12804, MobA-like NTP transferase (NTP_transf_3; gray, 32.9%); PF01467, Cytidylyltransferase-like (CTP_transf_like; orange, 23.6%); not visible are two genes matching PF02775 and PF02776 (both TPP-binding domains), which corresponds to 0.1% of non-redundant Ppm proteins. The blue slice represents 42 triple fusion proteins (2.8% of all Ppm proteins) with Ppm fused to both PF12804 and PF01467. c Distribution of genomic distances (in genes) between ppm and the nearest of either PF12804 or PF01467. Blue dots represent logarithm of the number of genes counted for each distance (corresponding to left y-axis), and the line represents the cumulative fraction of genomic distances (right y-axis); 59.3% of nearest nucleotidyltransferase genes (those closest to ppm) are fused to ppm (distance = 0), and only single counts occur at distances greater than ~350 genes. Inset: Number of genes for each distance up to 20, illustrating that two-thirds of nucleotidyltransferase genes are within ~5 genes of ppm