Fig. 5.
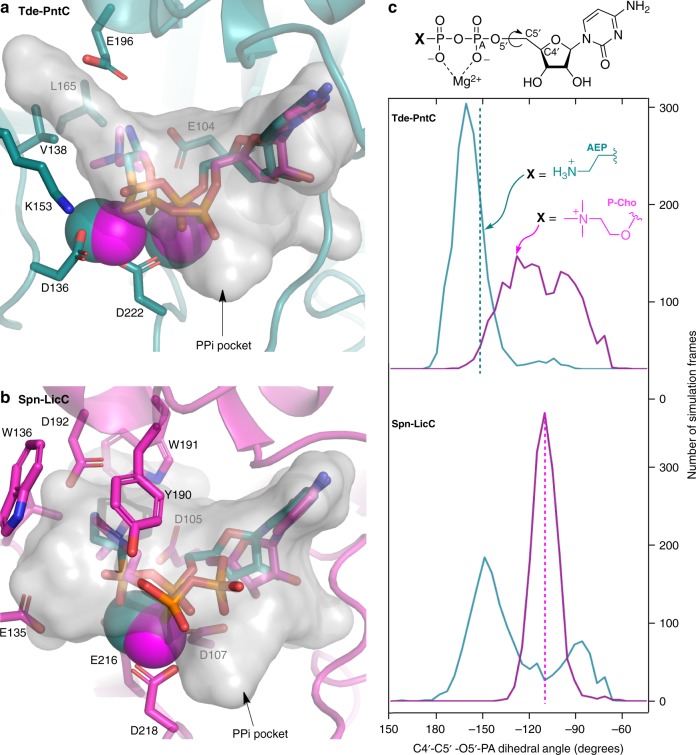
Molecular dynamics simulations of Tde-PntC (top) and Spn-LicC (bottom). a Representative conformations of the Tde-PntC active site with cognate ligand CMP-AEP and Mg2+ ions colored teal; non-cognate ligand CDP-Cho and Mg2+ ions are magenta. The transparent gray surface representation denotes the 287 Å3 cavity calculated from the crystal structure (minus CMP-AEP) using the Roll algorithm of POCASA42. b Representative conformations of the Spn-LicC active site showing the cognate ligand CDP-Cho and Mg2+ ion colored magenta; non-cognate ligand CMP-AEP and Mg2+ are colored teal. Transparent gray surface outlines the 381 Å3 cavity calculated from the 1JYL crystal structure coordinates after removing CDP-Cho (26). c Histogram of C4′-C5′-O5′-PA dihedral angles sampled during 10 × 150 ps independent simulations for each protein:ligand pair, using the same color scheme for each ligand (CMP-AEP = teal; CDP-Cho = magenta). Dotted lines represent crystallographically observed dihedral angles