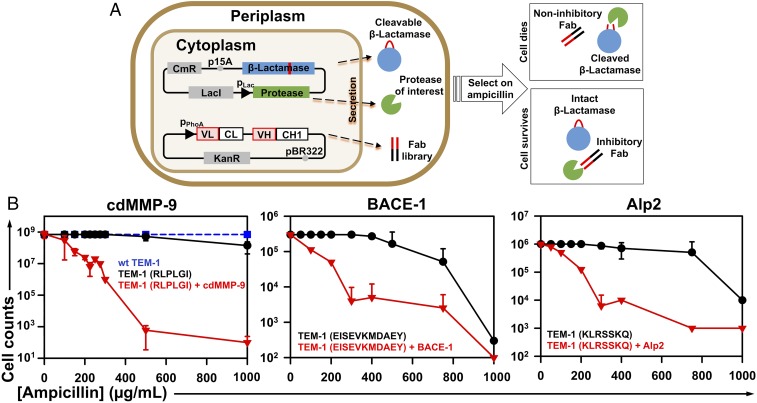
Fig. 1.
Functional selection design for protease inhibitory antibodies. (A) Scheme showing that 3 recombinant proteins are simultaneously coexpressed in the periplasmic space of E. coil—a clone from the Fab library, the protease of interest, and the modified β-lactamase TEM-1 with a cleavable peptide insertion. The protease extracellular/cd under a lac promoter and TEM-1 under its native promoter are cloned into a low copy number (p15A ori) plasmid of chloramphenicol resistance (CmR). The antibody Fab library under a phoA promoter is cloned into a medium copy number (pBR322 ori) plasmid carrying kanamycin resistance (KanR). If the Fab has no inhibition, the protease will cleave TEM-1 leading to cell death in the presence of ampicillin. An inhibitory Fab blocks proteolytic activity which allows TEM-1 to remain intact, resulting in cell growth on ampicillin plates. (B) Selection windows for cdMMP-9 (Left), BACE-1 (Middle), and Alp2 (Right) inhibitors. TEM-1 was modified by inserting the protease-specific cleavage peptide sequences (shown in parentheses) between Gly196 and Glu197 of TEM-1 (SI Appendix, Fig. S2). At 0 to 1,000 µg/mL ampicillin, survival curves of E. coli cells transformed with modified TEM-1s without protease genes were measured (black circles) and compared with those for cells coexpressing both modified TEM-1s and the associated proteases (red triangles). The survival curve with WT TEM-1 is shown as a blue dashed line. Experiments were repeated 3 times with 2×YT agar plates containing 0.1 mM IPTG.