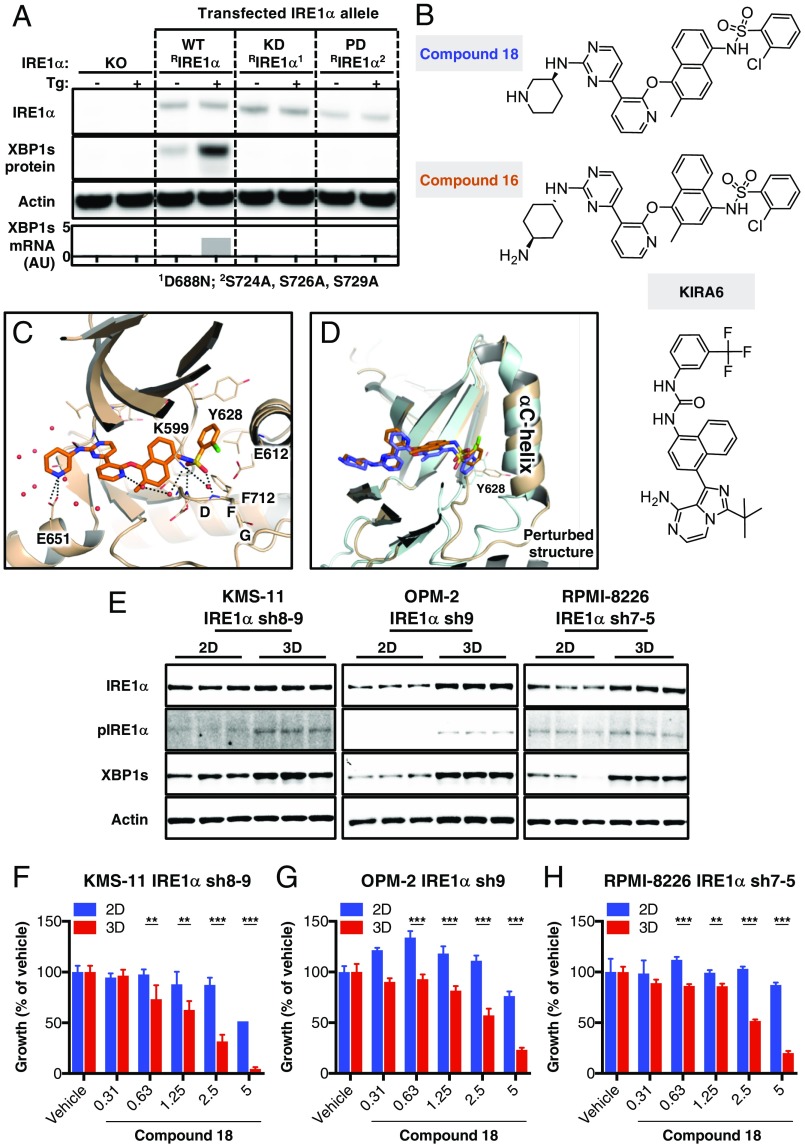
Fig. 3.
Importance of IRE1α kinase in RNase activation in growth of MM cell lines in 3D versus 2D. (A) KMS-11 IRE1α knockout (KO) cells (Cl. 2.3) were transiently transfected with expression plasmids encoding wild-type IRE1α (WT RIRE1α), a “kinase-dead” D688N mutant of IRE1α (KD RIRE1α), or an “autophosphorylation-deficient” S724A, S726A, S729A triple mutant of IRE1α (PD RIRE1α). Cells were then incubated in the absence or presence of thapsigargin (Tg, 100 nM) for 3 h and analyzed either by IB for levels of the indicated proteins or by RT-qPCR for mRNA levels of XBP1s. (B) Chemical structures of compounds 18 and 16 (35) and KIRA6 (42). (C) A close-up view of the crystal structure of 18 in complex with the kinase-RNase portion of IRE1α. The protein is rendered in ribbons with key residues in the ligand-binding pocket shown as sticks. Water molecules near the ligand are shown in red spheres. Black dashed lines indicate hydrogen bonding interactions. (D) Comparison between the cocrystal structures of 18 (colored in wheat and orange versus 16 (PDB ID code 4U6R, colored cyan and blue) bound to IRE1α. The C-terminal end of the αC-helix displays significant conformational changes between the two structures. (E) KMS-11, OPM-2, and RPMI-8226 cells were seeded on standard tissue culture plates (2D) or ULA plates followed by centrifugation to form single spheroids (3D). After 96 h, cells were lysed and analyzed by IB for indicated proteins. (F–H) Cells were seeded either in the 2D or 3D setting, treated for 150 h with vehicle (DMSO) or compound 18 at the indicated concentrations, and analyzed for cell growth by cell confluence using an Incucyte instrument (F) or cell viability using CellTiter-Glo 3D (G and H). **P ≤ 0.01, ***P ≤ 0.001.