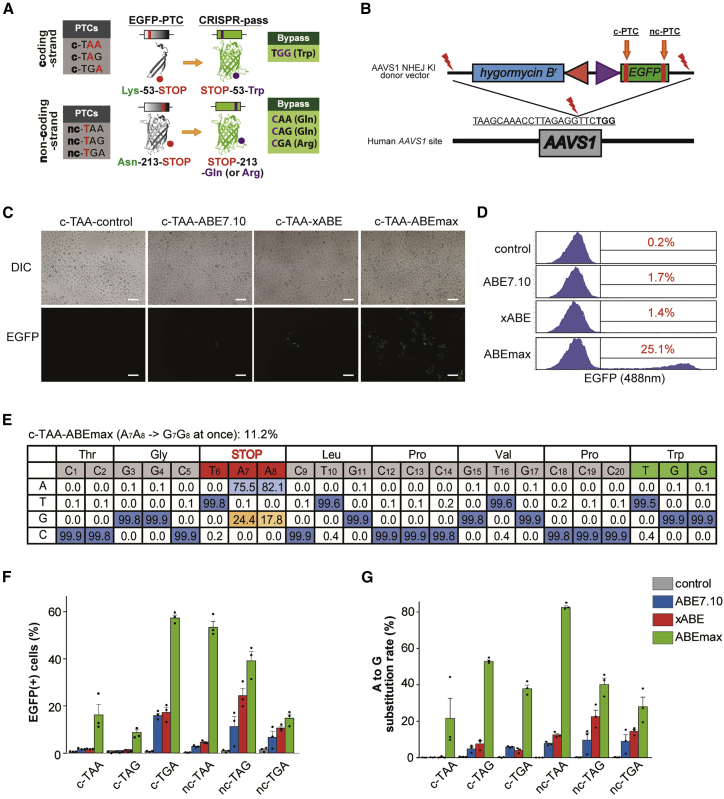
Figure 2.
Restoring the Function of EGFP Gene Expression in Six KI HeLa Cell Lines Carrying Various Types of PTCs
(A) Scheme for restoration of EGFP expression by CRISPR-pass. The first set of PTCs, which can be converted by targeting the coding strand, affect a residue that is located on a loop between the third and fourth beta strands; the second set of PTCs, which can be converted by targeting the noncoding strand, affect a residue that is located on a loop between the 10th and 11th beta strands. c-PTC, coding strand PTC; nc-PTC, noncoding strand PTC. GFP structures were originated from Wikimedia Commons created by Zephyris. (B) Schematic of NHEJ-mediated KI of the EGFP-PTC constructs into the AAVS1 site. Mutated EGFP KI cell lines were established for the three types of PTCs (TAA, TAG, and TGA). EGFP-PTC constructs were inserted into the AAVS1 site by NHEJ-mediated KI methods. The hygromysin B-resistant gene was also inserted for cell selection. (C) Fluorescence image of rescued EGFP expression in the c-TAA cell line after CRISPR-pass treatment. Three different versions of ABEs (ABE7.10, xABE, and ABEmax) were used for bypassing the PTCs in the EGFP gene. All scale bars are 100 μm. (D) Flow cytometry data after the different versions of ABEs (ABE7.10, xABE, and ABEmax) were treated in the c-TAA cell line. (E) Targeted deep-sequencing data showing the percentages of each of the four nucleotides at each position in the target DNA sequences as a substitution table, which was obtained from the c-TAA cell line after the ABEmax treatment. Bar graphs showing recovered EGFP expression levels as determined by flow cytometry (F) and showing A-to-G substitution rates at PTC sites as determined by targeted deep sequencing (G) for each EGFP-PTC KI cell line, after treatment with ABEs (ABE7.10, xABE, or ABEmax). Each dot represents the three independent experiments. Error bars represent SEM.