Figure 2. Standard RT-PCR using unmodified primers does not display strand-specificity.
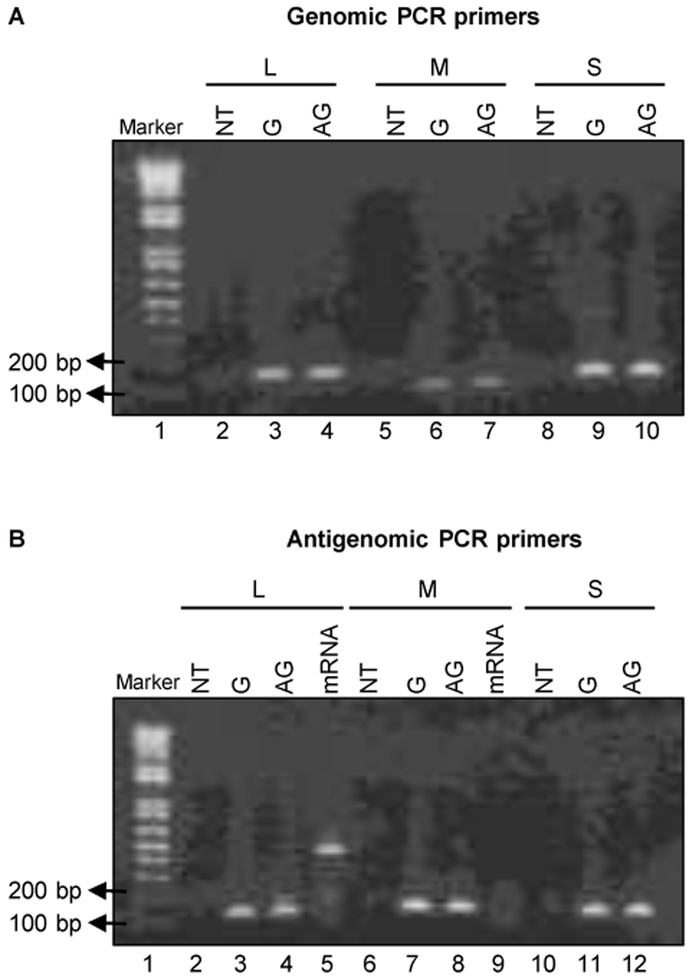
(A) 100 ng of in-vitro synthesized RNAs corresponding to the genomic (G) and antigenomic (AG) segments of L, M, and S RNAs were used for cDNA synthesis using unmodified RT primers specific for genomic L, M, and S segments respectively. The corresponding cDNAs were subjected to PCR by using unmodified PCR primer sets, specific for genomic L (lanes 2-4), M (lanes 5-7) and S (lanes 8-10) segments. NT represents the "no template" control for RT-PCR analysis (lanes 2, 5 and 8). (B) 100 ng of in-vitro synthesized RNAs corresponding to the genomic (G), antigenomic (AG) segments of L, M, and S RNAs, and cognate mRNAs (mRNA) of L and M segments were used for cDNA synthesis using unmodified RT primers specific for antigenomic L, M, and S segments respectively. The corresponding cDNAs were subjected to PCR by using an unmodified PCR primer set, specific for antigenomic L (lanes 2-5), M (lanes 6-9) and S (lanes 10-12) segments. Lane 1 in (A) and (B) represents the DNA size marker, and the location of the markers having 100 and 200 base pairs (bp) bands are indicated by arrows. The PCR products were analyzed by agarose gel electrophoresis. NT represents the no template control for RT-PCR analysis (lanes 2, 6 and 10).
