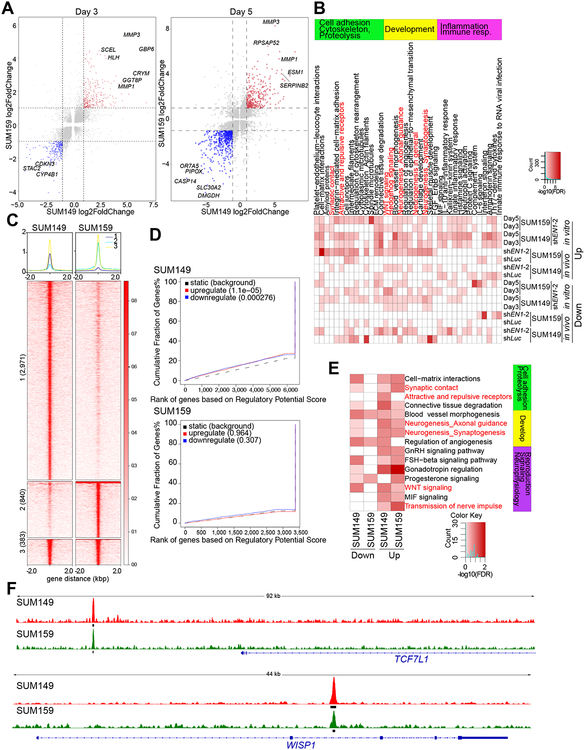
Figure 2. Transcriptional and genomic targets of EN1 in TNBC cells.
A, Gene expression changes in SUM149 (x-axis) and SUM159 (y-axis) cells expressing TET-inducible shEN1 grown in presence of doxycycline for 3 or 5 days. Red and blue indicates significantly up and downregulated genes, respectively, in both cell lines. B, Top process networks significantly enriched in genes differentially expressed following EN1 downregulation in SUM149 and SUM159 cell lines grown in cell culture (in vitro) or as xenografts (in vivo). C, Plot of ChIP-Seq signal of the EN1 peaks in SUM149 and SUM159 cells. Numbers indicate cell line-unique and overlapping peaks. D, BETA factor function prediction analysis to predict EN1 direct transcriptional function (activating/repressing) in SUM149 and SUM159 cells. Red and blue lines represent up and downregulated genes following EN1 knockdown. Black dashed line represents static genes. Genes are ranked by regulatory potential score, and significance relative to static genes is determined by Kolmogorov-Smirnov test. E, Top process networks significantly enriched in genes that are direct genomic targets of EN1 and are differentially expressed 3 days following EN1 downregulation in SUM149 and SUM159 cell lines. Each ChIP-Seq peak is assigned to the gene whose transcription start site (TSS) is closest to the peak, defining the factors direct genomic targets. F, EN1 ChIP-Seq signal at selected genes in SUM149 (red) and SUM159 (green) cells. Thick black lines below the signal tracks denote peaks.