Figure 1.
Analysis of Signal Peptide Encoding mRNAs Reveals that Secreted Proteins Tend to Escape Repression upon ER Stress
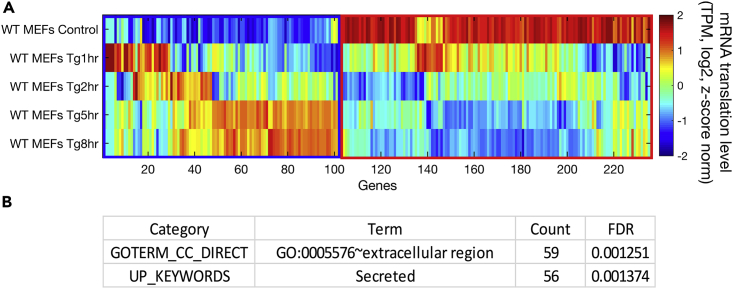
(A) Hierarchical clustering analysis was performed using 236 signal peptide-encoding mRNAs, with translation change of at least 2-fold compared with control in any of the time points examined following Tg treatment (1, 2, 5, and 8 h) in WT MEFs. The heatmap depicts hierarchal clustering of mRNA translation levels (transcripts per million [TPM] of ribosome footprint profiling in the respective samples, log2), according to Spearman's correlation. Z score normalization was further performed for visualization purposes. The mRNAs were clustered into two distinct groups: 102 induced mRNAs (blue square) and 134 repressed mRNAs (red square) following Tg treatment.
(B) Functional enrichment of induced signal peptide encoding genes was analyzed using DAVID (Huang da et al., 2009). The enriched terms were extracellular region and secreted proteins. False discovery rate (FDR)-corrected p values are indicated.