Figure 2.
Late-Specific ER Stress-Induced mRNAs Include Specific Amino Acid Biosynthesis Pathways and Their Cognate tRNA Synthetases
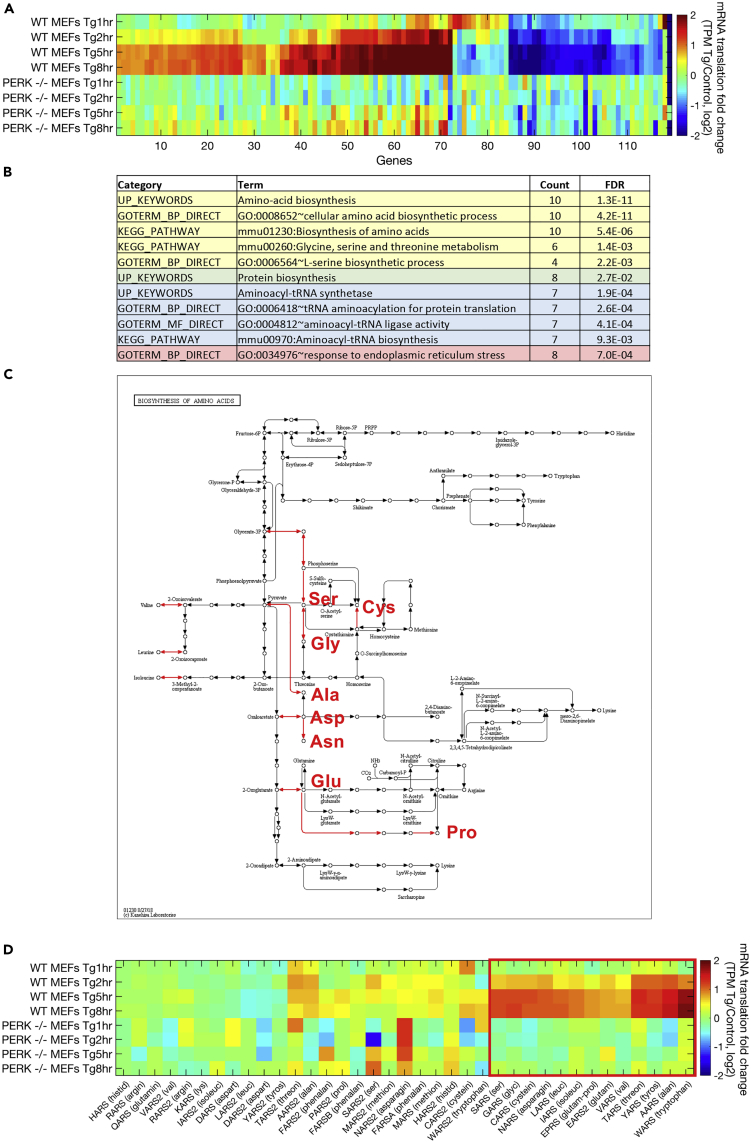
(A) Hierarchical clustering analysis was performed using 120 mRNAs that were differentially translated (identified using DESeq2, FDR-corrected p value < 0.1) between the group of 1- and 2-h ER-stressed WT MEFs and a second group of 5- and 8-h treated WT MEFs. The heatmap depicts hierarchically clustered log2 fold change in translation levels (transcripts per million [TPM] of ribosome footprint profiling in the respective samples, Tg/control, log2), according to Spearman's correlation. Clustering was performed according to WT MEF samples. As shown, PERK−/− cells display very little change in these mRNAs.
(B) Functional enrichment of the late-specific induced genes was analyzed using DAVID (Huang da et al., 2009). False discovery rate (FDR)-corrected p values are indicated.
(C) An amino acid biosynthesis metabolic map that was generated using the KEGG mapper (Kanehisa and Goto, 2000) of late-specific ER stress-induced genes shows induction of biosynthesis pathways (induced reaction arrows are colored in red) for eight non- or partly essential amino acids: Ser, Cys, Gly, Ala, Asp, Asn, Glu, and Pro.
(D) Hierarchical clustering analysis of translation log2 fold changes (as described in A) for the group of all tRNA synthetases, identified a cluster of ER stress-induced, PERK-dependent, tRNA synthetases (marked by a red square).