Abstract
Introduction
Molecular pathways involved in ligamentum flavum (LF) hypertrophy are still unclarified. The purpose of this study was to characterize LF hypertrophy by microRNA (miRNA) profiling according to the classification of lumbar spinal stenosis (LSS).
Methods
Classification of patients with LSS into ligamentous and non-ligamentous cases was conducted by clinical observation and the morphometric parameter adopting the LF/spinal canal area ratio (LSAR) from measurements of magnetic resonance imaging (MRI) T2 weighed images. LF from patients with ligamentous stenosis (n=10) were considered as the degenerative hypertrophied samples, and those from patients with non-ligamentous LSS (n=7) and lumbar disc herniation (LDH, n=3) were used as non-hypertrophied controls. Profiling of miRNA from all samples was conducted by Agilent microarray. Microarray data analysis was performed with GeneSpring GX, and pathway analysis was performed using Ingenuity Pathway Analysis.
Results
The mean LSAR in the ligamentous group was significantly higher than that in the control group (0.662±0.154 vs 0.301±0.068, p=0.0000171). Ten significantly differentially expressed miRNA were identified and taken as a signature of LF hypertrophy: nine miRNA showed down-regulated expression, and one showed up-regulated expression in the ligamentous LF. Among those, miR-423-5p (rs=-0.473, p<0.05), miR-4306 (rs=-0.628, p<0.01), miR-516b-5p (rs=-0.629, p<0.01), and miR-497-5p (rs=0.461, p<0.05) were correlated to the LSAR. Pathway analysis predicted aryl hydrocarbon receptor signaling (p<0.01), Wnt/β-catenin signaling (p<0.01), and insulin receptor signaling (p<0.05) as canonical pathways associated with the miRNA signature.
Conclusions
Classification based on quantification of the MRI axial image is useful for studying hypertrophy of the LF. Aryl hydrocarbon receptor and Wnt/β-catenin signaling may be involved in LF hypertrophy.
Keywords: lumbar spinal stenosis, ligamentum flavum, miRNA, hypertrophy
Introduction
Lumbar spinal stenosis (LSS), the most common spinal disorder in elderly people, causes lower back pain and walking disability with intermittent claudication1,2). Yet, recent guidelines describe LSS as syndromic, and its etiology is still unclear, although it can be classified into distinct diseases (Diagnosis and treatment of degenerative lumbar spinal stenosis in Evidence-based clinical guidelines for multidisciplinary spine care by the North American Spine Society, 2011; Clinical practice guideline on the diagnosis and treatment of lumbar spinal stenosis by the Japanese Orthopaedic Association, 2011). Although multiple factors have been suggested to be involved in the development of LSS, hypertrophy of the ligamentum flavum (LF), which connects the vertebra and covers the dorsal walls of the spinal canals, narrows the spinal canal space consequently impinging the nerves, and this has been considered as a cause of LSS. The pathology of degenerative hypertrophy is characterized by loss of elastic fibers and the appearance of a focal disorganized collagenous matrix, which is often associated with chondrometaplasia3,4). Although several cytokines such as TGFβ, CTGF, FGF2, and Angptl2 have been identified to be involved in LF hypertrophy, the molecular mechanism underlying the hypertrophy still remains largely unknown3,5-9).
MicroRNAs (miRNAs) are short non-coding RNAs that predominantly bind to the coding or 3'-untranslated region of the target mRNA to inhibit translation by promoting the degradation and/or blocking of the translational complex of the target mRNA, thereby playing important roles as post-transcriptional regulators10,11). MiRNAs potentially regulate multiple mRNA targets and have been reported to be involved in various physiological and pathological conditions10). Interestingly, the products of multiple targets often belong to the molecules of specific signaling pathways11). Therefore, pathway analysis of the miRNA transcriptome has been applied to elucidate the pathways playing important roles in various biological and pathogenic situations12).
It has been recently reported that there is a difference in the clinical outcome between cases of ligamentous and non-ligamentous stenosis based on a novel classification criterion according to the measurement of magnetic resonance imaging (MRI) T2 axial images of the lumbar spine13). The classification adopts information from clinical evaluations and the morphometric analysis of MRI images, especially the ratio of the cross sectional area (CSA) of the LF to that of the spinal canal space (denoted as the LSAR hereafter). Notably, ligamentous stenosis in patients with LSS showed better outcomes from conservative treatment13). Thus, classification by the area ratio, rather than the un-normalized actual thickness measurements, may also be applied to evaluate the potentially pathogenic hypertrophy of the LF in LSS.
To elucidate the molecular pathways involved in LF hypertrophy, we conducted profiling of the miRNAs expressed in the LF from patients that were sorted according to this new morphometric-based classification.
Materials and Methods
Subjects
LF specimens were surgically obtained from 20 patients with LSS or lumbar disc herniation (LDH) between 2010 and 2013. Diagnosis was made by two spinal surgeons (Y.S. and A.H). Classification of non-ligamentous and ligamentous stenosis was done as described by Sakai et al.13), adopting LSAR from measurements of MRI T2-weighted images. This classification can define hypertrophy of the LF, which is a main cause of stenosis and the resulting clinical symptoms. The LF samples from the most severely stenosed parts of the patients with ligamentous LSS (n=10) were taken as hypertrophied LF samples, and those obtained during the spinal surgery of the patients with non-ligamentous LSS (n=7) and LDH (n=3) were taken as non-hypertrophied controls. This comparison allowed us to focus specifically on extraction of the pathways involved in LF hypertrophy rather than simplify classifying these pathways for LSS generally. LF samples were obtained from the most severely stenosed joints in patients with LSS or from the surgical joints in patients with LDH. The subjects' age, gender, and LSAR at the joint where the LF was analyzed are listed in Table 1 and are summarized in Table 2. There is no statistical difference in the mean ages between ligamentous and non-ligamentous groups (75.70±4.83 and 70.70±10.98, respectively). All of the LF specimens and clinical information from the patients were obtained after receiving informed consent. The study was approved by the Institutional Review Board of the ethics and conflicts of interest committee.
Table 1.
Sample Information.
| Sample ID |
Age | Sex | Diagnosis | Level* | LSAR** |
|---|---|---|---|---|---|
| L1 | 74 | Female | Lumbar spinal stenosis | L4-5 | 0.67 |
| L2 | 72 | Female | Lumbar spinal stenosis | L4-5 | 0.49 |
| L3 | 80 | Male | Lumbar spinal stenosis | L4-5 | 0.49 |
| L4 | 70 | Male | Lumbar spinal stenosis | L4-5 | 0.53 |
| L5 | 79 | Female | Lumbar spinal stenosis | L3-4 | 0.59 |
| L6 | 79 | Female | Lumbar spinal stenosis | L4-5 | 0.80 |
| L7 | 67 | Male | Lumbar spinal stenosis | L3-4 | 0.87 |
| L8 | 82 | Male | Lumbar spinal stenosis | L3-4 | 0.74 |
| L9 | 78 | Male | Lumbar spinal stenosis | L4-5 | 0.89 |
| L10 | 76 | Male | Lumbar spinal stenosis | L5-S1 | 0.55 |
| N1 | 73 | Male | Lumbar spinal stenosis | L3-4 | 0.30 |
| N2 | 91 | Female | Lumbar spinal stenosis | L4-5 | 0.38 |
| N3 | 74 | Male | Lumbar spinal stenosis | L4-5 | 0.33 |
| N4 | 64 | Female | Lumbar disc herniation | L4-5 | 0.15 |
| N5 | 57 | Female | Lumbar disc herniation | L2-3 | 0.27 |
| N6 | 63 | Male | Lumbar disc herniation | L3-4 | 0.34 |
| N7 | 87 | Female | Lumbar spinal stenosis | L2-3 | 0.36 |
| N8 | 62 | Female | Lumbar spinal stenosis | L4-5 | 0.23 |
| N9 | 66 | Male | Lumbar spinal stenosis | L4-5 | 0.32 |
| N10 | 70 | Male | Lumbar spinal stenosis | L4-5 | 0.33 |
*The levels of the spinal joints where ligamentum flavum (LF) samples were obtained.
**The ratio of cross sectional area of LF to that of spinal canal in the joint where LF samples were obtained.
Table 2.
Sample Summary.
| Control (non-ligamentous) | Ligamentous | p value | |
|---|---|---|---|
| Case (n) | 10 | 10 | |
| Diagnosis* | Lumbar spinal stenosis (7) | Lumbar spinal stenosis (10) | |
| Lumbar disc herniation (3) | |||
| Age (yrs) [range] | 70.70±10.98[57-91] | 75.70±4.83[67-82] | 0.211 |
| Sex (M:F) | 5:5 | 6:4 | 0.653** |
| CSA of LF (mm2) | 60.26±16.71 | 121.90±31.91 | 0.000102 |
| CSA of spinal canal (mm2) | 200.04±28.26 | 188.22±42.66 | 0.479 |
| LSAR*** | 0.301±0.068 | 0.662±0.154 | 0.0000171 |
*Parentheses, number of cases
**Fisher’s exact test
*** Ligamentum flavum/spinal canal area ratio
Data are expressed as mean±SD.
Tissue and RNA preparation
All specimens were immediately washed with ice-cold phosphate-buffered saline after surgical removal, stored in a liquid nitrogen tank, and then kept at -80°C until the experiments were performed. The tissue samples included full layers of LF. Each frozen LF tissue (100-300 mg) in a 13-mL master aluminum case with 15-mm stainless beads was homogenized using Shake Master NEO (Bio Medical Sciences, Tokyo, Japan) at 1,500 rpm for 2 min under liquid nitrogen twice. The total RNA, including miRNA of the LF homogenates, was isolated using a miRNeasy kit (QIAGEN, Hilden, Germany), according to the manufacturer's instructions. The total RNA was treated with RNase-free DNase (QIAGEN) to eliminate possible contamination with genomic DNA. The total RNA was eluted in 200 μL of nuclease-free water from four silica columns, concentrated to 25 μL using RNeasy MinElute Cleanup Kit (QIAGEN), and stored at -80°C for further analysis. The quality of total RNA was checked using an Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
miRNA array
One hundred nanograms of the isolated RNA from each sample was labeled using miRNA Complete Labelling and Hyb Kit (Agilent Technologies) according to the manufacturer's protocols. The labeled RNA was hybridized to SurePrint G3 Human miRNA Microarray (2,588 miRBase mature miRNAs, Release 21.0, Agilent Technologies). The microarray was scanned using one color scan setting for 8X 60K array slides with a G2505C Agilent DNA microarray scanner. Images were extracted with Feature Extraction Software 10.7.3.1 (Agilent Technologies).
Data analysis
The gene expression data were normalized by per-chip normalization and per-gene normalization using GeneSpring GX (Agilent Technologies). For per-chip normalization, all expression data on a chip were normalized to the 25th and 75th percentiles of all values on that chip. For per-gene normalization, the data for a given gene were normalized to the median expression level of that gene across all samples. The normalized data were subjected to statistical analysis for comparisons between the control (non-ligamentous, n=10) and hypertrophied LF (ligamentous, n=10) groups, and the differentially expressed entities were extracted by Welch's t-test (cutoff, p<0.05). To extract the entities that exhibited an age- or LSAR-related pattern, correlation analysis using all samples analyzed (n=20) was performed by Spearman's rank correlation test (cutoff 0.4<|rs|) with Bonferroni correction using GeneSpring software. In this study, Spearman's rank correlation test was applied for all correlation analyses, unless otherwise noted. For pathway analysis, the targets of the differentially expressed miRNAs were extracted by GeneSpring according to TargetScan (cutoff, p<0,01). The targets (483 genes) were analyzed with Ingenuity Pathway Analysis (IPA, QIAGEN). The p-values of the pathway analysis were calculated by IPA.
Results
Ratio of LF to spinal canal CSAs
As shown in Table 2, the mean CSA of the LF, but not the mean CSA of the spinal canal, at the most severely stenosed part of the spine was significantly increased in the ligamentous stenosis group (121.90±31.91 vs 60.26±16.71, p=0.000102). The LSAR was significantly higher in the ligamentous stenosis group (0.662±0.154 vs 0.301±0.068, p=0.0000171). The age difference between the groups was not statistically significant (75.70±4.83 vs 70.70±10.98, p=0.211).
Age-related expression of miRNA
The miRNAs with expression levels that were significantly correlated to donor age were miR-29c-3p (rs=0.561, p<0.05), miR-595 (rs=0.507, p<0.05), miR-663b (rs=0.563, p<0.05), miR-1290 (rs=-0.543, p<0.05), and miR-223-3p (rs=0.463, p<0.05).
miRNAs correlated to the area ratio
Differentially expressed miRNAs between the hypertrophied LF and control LF groups were extracted and are shown in Table 3. Ten miRNAs showed significantly altered expression between groups (p<0.05): the expression levels of nine miRNA were decreased in the hypertrophied ligaments, and the expression level of one miRNA was increased. The heatmap of the expression values is shown in Fig. 1. The Spearman's rank test was applied for correlation analysis to the LSAR. Among the differentially expressed miRNAs (Table 3), miR-423-5p (rs=-0.473, p<0.05), miR-4306 (rs=-0.628, p<0.01), miR-516b-5p (rs=-0.629, p<0.01), and miR-497-5p (rs=0.461, p<0.05) were also correlated to the LSAR.
Table 3.
Differentially Expressed miRNAs.
| miRNA species | Regulation | Fold Change | p value |
|---|---|---|---|
| miR-1228-3p | Down | -1.26 | 0.04318 |
| miR-1237 | Down | -5.38 | 0.02298 |
| miR-30c-2-3p | Down | -1.21 | 0.02649 |
| miR-423-5p | Down | -1.18 | 0.00510 |
| miR-4306 | Down | -1.26 | 0.00464 |
| miR-483-5p | Down | -1.31 | 0.02848 |
| miR-497-5p | Up | 1.51 | 0.03140 |
| miR-514b-5p | Down | -1.24 | 0.02212 |
| miR-516b-5p | Down | -5.26 | 0.00392 |
| miR-765 | Down | -1.36 | 0.00950 |
Regulation and fold change indicate the comparison of ligamentous group to control group.
Figure 1.
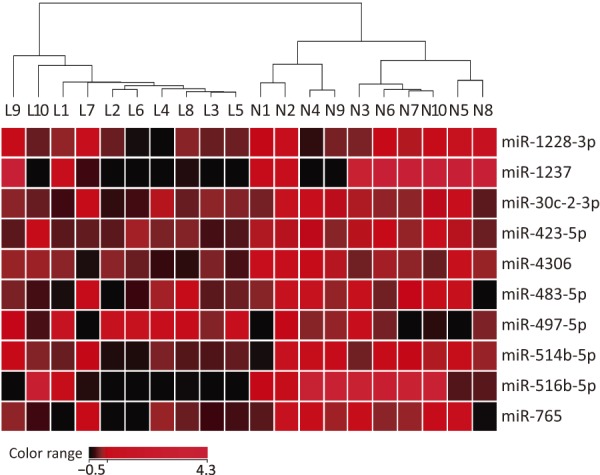
Heatmap of the differentially expressed miRNA. The sample names (Ligamentous group, L1-L10; control group, N1-N10) correspond to Table 1. The hierarchical clustering of the samples was conducted by Euclidean measurement and Ward’s method for linkage using normalized intensity values.
Pathway analysis
The signature of the miRNAs for ligament hypertrophy, listed in Table 3, was applied to pathway analysis to identify the molecular networks involved in LF hypertrophy. The canonical pathways were predicted as those involved in LF hypertrophy (Table 4); four pathways were statistically significant (p<0.01). Among them, aryl hydrocarbon receptor (AHR) signaling and Wnt/β-catenin signaling were shown as networks with interacting molecules involved in the pathways, and the miRNA signatures were extracted, as shown in Fig. 2. Insulin receptor signaling was also extracted as a canonical pathway with mild significance (p<0.05).
Table 4.
Predicted Pathways Involving the miRNA.
| Canonical pathways | p value |
|---|---|
| Aryl hydrocarbon receptor signaling | 0.000776 |
| Pentose phosphate pathway (oxidative branch) | 0.00278 |
| Wnt/β-catenin signaling | 0.00361 |
| GADD45 signaling | 0.00774 |
| Insulin receptor signaling | 0.0106 |
Figure 2.
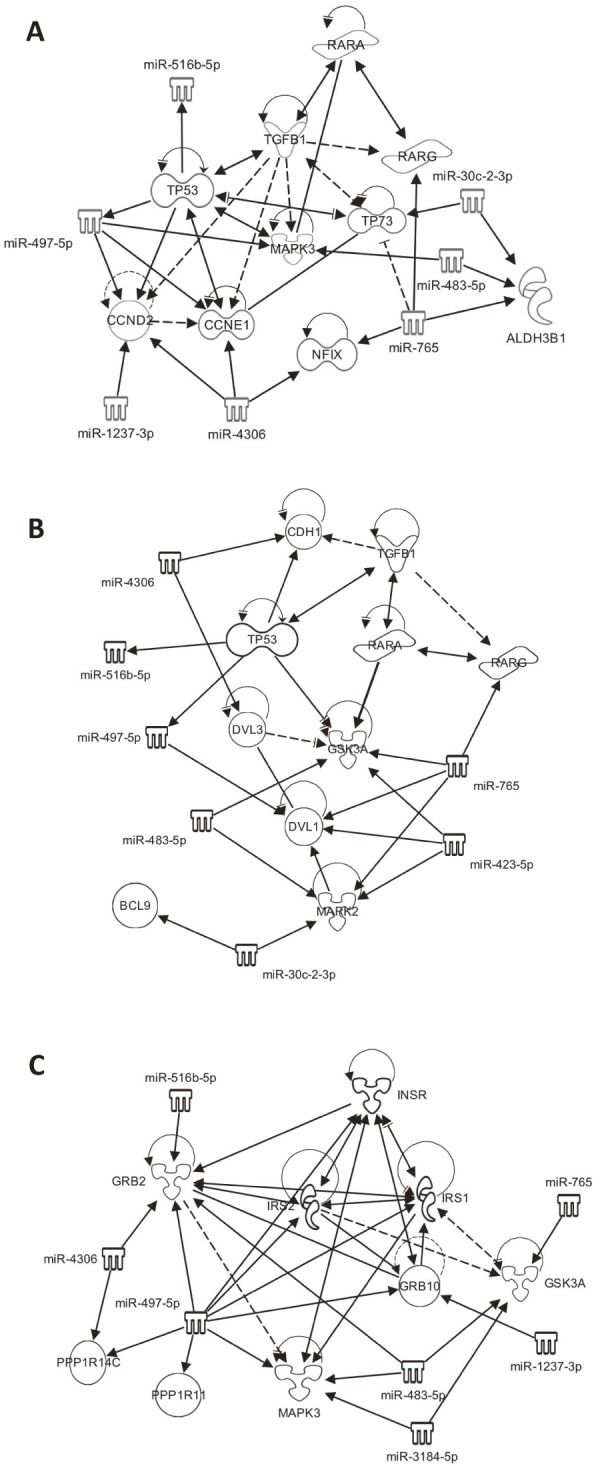
Canonical pathways involved in LF hypertrophy predicted by miRNA signature. A, AHR signaling. B, Wnt/β-catenin signaling. C, Insulin receptor signaling. The estimated network through IPA on the target genes of the miRNA signature for LF hypertrophy listed in Table 3. Genes/miRNA directly (solid arrow) and indirectly (broken arrow) interacted with them.
Discussion
The number of comprehensive ‘omics’ studies to elucidate the molecular pathology of LF degenerative hypertrophy in LSS patients has been increasing7,14,15). In these cases, LF samples from LDH patients, for which the age group is often younger than that of patients with LSS, were used as controls. Several studies have been reported that the thickness of the LF is associated with age16-19). In fact, the area ratio was associated to age (rs=0.548, p=0.012) in the present subjects (n=20), and age-related miRNA expression patterns were also identified in this study. Using the novel classification system proposed by Sakai et al., the pathogenic LF hypertrophy among LSS patients could be selected by defining ligamentous stenosis13). Moreover, the non-hypertrophied LF could also be obtained from elderly patients of a similar age group to the LSS group with pathogenic LF hypertrophy. In addition to the diagnostic significance of the classification13), the morphometric parameter LSAR is a candidate quantitative clinical marker for molecular profiling to study the molecular mechanism of LF hypertrophy.
The age-related miRNAs were extracted with mild significance (p<0.05), and differentially expressed miRNAs between patients with hypertrophied and non-hypertrophied LF were identified. These miRNAs were not overlapping, suggesting that age-related thickening and pathological hypertrophy of the LF may have distinct molecular pathologies. Recently, the miRNA species involved in LF hypertrophy have been reported. Chen et al. compared LSS and LDH samples and showed that the level of miR-155 in the LF was associated with LF thickness (r=0.958, p<0.01), and the mRNA and protein expression levels of type I and III collagen were increased20). Xu et al. employed a microarray technique to samples from patients with LSS and LDH, miR-221, whose expression level was increased in the LF from patients with LSS, regulates LF hypertrophy by suppressing TIMP2, which inhibits the degradation of type I and III collagen, and consequently increases the amount of these collagens7). In this study, neither these miRNAs were extracted as those significantly related to LF hypertrophy; however, miR-29c and miR-223, which were identified as age-related miRNAs in this study, were also listed as differentially expressed miRNAs by Xu et al7). This difference is possibly due to the selection of control samples, but further analysis is required for a detailed explanation.
It is widely accepted that fibrotic alteration is a major event in the development of LF hypertrophy3). Only miR-497-5p was identified to be increased in the hypertrophied LF and was correlated positively to the LSAR. It was recently described that miR-497-5p induced myofibroblast differentiation of lung resident mesenchymal stem cells resulting in pulmonary fibrosis21). Cells with a myofibroblastic nature in the LF may be involved in the hypertrophy22), thus suggesting that miR-497-5p possibly plays a role in hypertrophy via the promotion of myofibroblast in LF. It has been reported that miR-4306 was significantly decreased in the sclerotic samples of patients with arteriosclerosis obliterans, characterized by fibrosis of the tunica intima23). Thus, these miRNAs may also play a role in the fibrotic change of LF. Although other miRNAs, miR-423-5p and miR516b-5p, which were also extracted as those expressed related to LSAR, are potentially involved in the degenerative hypertrophy, the functional information related to the pathology have not been available at this time. Further functional analyses of these four miRNAs are required to demonstrate direct implication in LF hypertrophy.
The canonical pathways that these miRNAs are potentially involved in were predicted by IPA. AHR signaling is triggered by the activation of AHR, which is an intracellular receptor-type transcription factor responsible for the induction of enzymes in drug metabolism and is involved in immune responses and cell cycle control24,25). Among the major ligands for AHR is tetrachlorodibenzodioxin (TCDD), which is elevated in the serum by smoking. Ligand-evoked AHR signaling modulates inflammation and is possibly associated with atherosclerosis development26,27). In addition, ligand-independent action of AHR signaling has also been demonstrated, and thus the regulatory function of AHR signaling is widespread in various biological and pathological conditions28,29). Ahr knockout mice exhibited liver fibrosis and ligand-independent down-regulation of TGFβ1 expression is thought to be involved in the pathogenesis30,31). Wnt/β-catenin signaling also plays pleiotropic roles in developmental, physiological, and pathological situations; e.g., regulation of the bone accrual and integrity of the articular joints32,33). Cyclic strain induced β-catenin signaling has been reported in cultured LF cells from patient with ossification of the LF34). Although it was less statistically significant, insulin receptor signaling was extracted as the hypertrophy-related pathway, suggesting that the alteration of the signaling, such as insulin resistance, which is often associated with common diseases and/or age-related disorders, may contribute to hypertrophy35). Pentose phosphate and GADD45 signaling pathways have been described to be involved in oxidative stress response and inflammation36,37). These pathways have not been described in the context of LF hypertrophy but are attractive points of views from which to study LF hypertrophy.
Interestingly, TGFβ1 is involved in both the AHR and Wnt/β-catenin pathways38-41). TGFβ is known as a major regulator of tissue fibrosis and has also been suggested to play important roles in LF hypertrophy42). In general, tissue fibrosis is a complex consequence of inflammation42). Although the age-related decline of tissue homeostasis, such as the loss of elastic fibers in the LF, showed fibrotic alteration, it is suggested that inflammation-induced TGFβ signaling acts specifically on hypertrophy3,43). Moreover, it is known that senescent cells are present in most aged tissues, and the secretion of pro-inflammatory cytokines and chemokines is a characteristic of senescent cells, representing the so-called senescence associated secretory phenotype44,45). Saito et al. recently demonstrated that both mechanical stress and macrophage infiltration are involved in the development of severe hypertrophy in the LF in a mouse model43). Long-term mechanical stress reduced the integrity of elastic fibers in the mouse LF with no substantial increase of TGFβ1 expression; however, macrophage infiltration could induce TGFβ1 expression in the LF tissues. It is therefore conceivable that the age-related histological alteration of the LF and the signals or events to directly promote the hypertrophy may be distinct mechanisms that are nevertheless collaboratively involved in LF hypertrophy.
The main limitations of the present study are centered on the minimal number of samples used for profiling and a lack of the validation by histological analyses. The pathology of the hypertrophied LF is not uniform but rather a complex pathology of the degenerative changes, such as loss of elastic fibers, accumulation of a collagenous matrix, increased vascularization, and appearance of chondrometaplasia3). Increasing the number of the samples or adopting a focal approach such as microdissection-based sampling, as well as manipulation into useful models, may be required for further validation. Nevertheless, this study provides new insight into the morphometric parameter and potential pathways that may regulate hypertrophy of the LF.
Conflicts of Interest: The authors declare that there are no conflicts of interest.
Sources of funding: This study was supported by the Research Funding for Longevity Sciences (24-12 and 27-16) from NCGG and the Program for Promotion of Fundamental Studies in Health Sciences conducted by the National Institute of Biomedical Innovation of Japan (10-43).
Acknowledgements
The authors would like to thank the participants who donated LF samples and Biobank, NCGG.
References
- 1.Katz JN, Harris MB. Clinical practice. Lumbar spinal stenosis. The New England JOurnal of Medicine. 2008;358(8):818-25. [DOI] [PubMed] [Google Scholar]
- 2.Siebert E, Pruss H, Klingebiel R, et al. Lumbar spinal stenosis: syndrome, diagnostics and treatment. Nature Reviews Neurology. 2009;5(7):392-403. [DOI] [PubMed] [Google Scholar]
- 3.Sairyo K, Biyani A, Goel V, et al. Pathomechanism of ligamentum flavum hypertrophy: a multidisciplinary investigation based on clinical, biomechanical, histologic, and biologic assessments. Spine. 2005;30(23):2649-56. [DOI] [PubMed] [Google Scholar]
- 4.Schrader PK, Grob D, Rahn BA, et al. Histology of the ligamentum flavum in patients with degenerative lumbar spinal stenosis. European spine journal: official publication of the European Spine Society, the European Spinal Deformity Society, and the European Section of the Cervical Spine Research Society. 1999;8(4):323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura T, Okada T, Endo M, et al. Angiopoietin-like protein 2 induced by mechanical stress accelerates degeneration and hypertrophy of the ligamentum flavum in lumbar spinal canal stenosis. PloS One. 2014;9(1):e85542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T, Okada T, Endo M, et al. Angiopoietin-like protein 2 promotes inflammatory conditions in the ligamentum flavum in the pathogenesis of lumbar spinal canal stenosis by activating interleukin-6 expression. European spine journal: official publication of the European Spine Society, the European Spinal Deformity Society, and the European Section of the Cervical Spine Research Society. 2015;24(9):2001-9. [DOI] [PubMed] [Google Scholar]
- 7.Xu YQ, Zhang ZH, Zheng YF, et al. MicroRNA-221 regulates hypertrophy of ligamentum flavum in lumbar spinal stenosis by targeting TIMP-2. Spine. 2015. [DOI] [PubMed] [Google Scholar]
- 8.Honsawek S, Poonpukdee J, Chalermpanpipat C, et al. Hypertrophy of the ligamentum flavum in lumbar spinal canal stenosis is associated with increased bFGF expression. International Orthopaedics. 2013;37(7):1387-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong ZM, Zha DS, Xiao WD, et al. Hypertrophy of ligamentum flavum in lumbar spine stenosis associated with the increased expression of connective tissue growth factor. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2011;29(10):1592-7. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai Y, Ito S, Hida T, et al. Clinical outcome of lumbar spinal stenosis based on new classification according to hypertrophied ligamentum flavum. Journal of orthopaedic science: official journal of the Japanese Orthopaedic Association. 2017;22(1):27-33. [DOI] [PubMed] [Google Scholar]
- 14.Kamita M, Mori T, Sakai Y, et al. Proteomic analysis of ligamentum flavum from patients with lumbar spinal stenosis. Proteomics. 2015;15(9):1622-30. [DOI] [PubMed] [Google Scholar]
- 15.Yabe Y, Hagiwara Y, Ando A, et al. Chondrogenic and fibrotic process in the ligamentum flavum of patients with lumbar spinal canal stenosis. Spine. 2015;40(7):429-35. [DOI] [PubMed] [Google Scholar]
- 16.Abbas J, Hamoud K, Masharawi YM, et al. Ligamentum flavum thickness in normal and stenotic lumbar spines. Spine. 2010;35(12):1225-30. [DOI] [PubMed] [Google Scholar]
- 17.Kolte VS, Khambatta S, Ambiye MV. Thickness of the ligamentum flavum: correlation with age and its asymmetry-an magnetic resonance imaging study. Asian Spine Journal. 2015;9(2):245-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munns JJ, Lee JY, Espinoza Orias AA, et al. Ligamentum flavum hypertrophy in asymptomatic and chronic low back pain subjects. PloS One. 2015;10(5):e0128321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Safak AA, Is M, Sevinc O, et al. The thickness of the ligamentum flavum in relation to age and gender. Clinical Anatomy. 2010;23(1):79-83. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Liu Z, Zhong G, et al. Hypertrophy of ligamentum flavum in lumbar spine stenosis is associated with increased miR-155 level. Disease markers. 2014;2014:786543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Shi C, Wang C, et al. The role of miR-497-5p in myofibroblast differentiation of LR-MSCs and pulmonary fibrogenesis. Scientific Reports. 2017;7:40958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hur JW, Bae T, Ye S, et al. Myofibroblast in the ligamentum flavum hypertrophic activity. European spine journal: official publication of the European Spine Society, the European Spinal Deformity Society, and the European Section of the Cervical Spine Research Society. 2017. [DOI] [PubMed] [Google Scholar]
- 23.He XM, Zheng YQ, Liu SZ, et al. Altered Plasma MicroRNAs as Novel Biomarkers for Arteriosclerosis Obliterans. Journal of Atherosclerosis and Thrombosis. 2016;23(2):196-206. [DOI] [PubMed] [Google Scholar]
- 24.Schrenk D. Impact of dioxin-type induction of drug-metabolizing enzymes on the metabolism of endo- and xenobiotics. Biochemical Pharmacology. 1998;55(8):1155-62. [DOI] [PubMed] [Google Scholar]
- 25.Mimura J, Ema M, Sogawa K, et al. Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes & Development. 1999;13(1):20-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu D, Nishimura N, Kuo V, et al. Activation of aryl hydrocarbon receptor induces vascular inflammation and promotes atherosclerosis in apolipoprotein E-/- mice. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31(6):1260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor is activated by modified low-density lipoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(4):1412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn ME, Allan LL, Sherr DH. Regulation of constitutive and inducible AHR signaling: complex interactions involving the AHR repressor. Biochemical Pharmacology. 2009;77(4):485-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stockinger B, Di Meglio P, Gialitakis M, et al. The aryl hydrocarbon receptor: multitasking in the immune system. Annual Review of Immunology. 2014;32:403-32. [DOI] [PubMed] [Google Scholar]
- 30.Chang X, Fan Y, Karyala S, et al. Ligand-independent regulation of transforming growth factor beta1 expression and cell cycle progression by the aryl hydrocarbon receptor. Molecular and Cellular Biology. 2007;27(17):6127-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Salguero P, Pineau T, Hilbert DM, et al. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268(5211):722-6. [DOI] [PubMed] [Google Scholar]
- 32.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nature Medicine. 2013;19(2):179-92. [DOI] [PubMed] [Google Scholar]
- 33.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192-205. [DOI] [PubMed] [Google Scholar]
- 34.Cai HX, Yayama T, Uchida K, et al. Cyclic tensile strain facilitates the ossification of ligamentum flavum through beta-catenin signaling pathway: in vitro analysis. Spine. 2012;37(11):E639-46. [DOI] [PubMed] [Google Scholar]
- 35.Facchini FS, Hua N, Abbasi F, et al. Insulin resistance as a predictor of age-related diseases. The Journal of Clinical Endocrinology and Metabolism. 2001;86(8):3574-8. [DOI] [PubMed] [Google Scholar]
- 36.de la Fuente H, Cibrian D, Sanchez-Madrid F. Immunoregulatory molecules are master regulators of inflammation during the immune response. FEBS Letters. 2012;586(18):2897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perl A, Hanczko R, Telarico T, et al. Oxidative stress, inflammation and carcinogenesis are controlled through the pentose phosphate pathway by transaldolase. Trends in Molecular Medicine. 2011;17(7):395-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silginer M, Burghardt I, Gramatzki D, et al. The aryl hydrocarbon receptor links integrin signaling to the TGF-beta pathway. Oncogene. 2016;35(25):3260-71. [DOI] [PubMed] [Google Scholar]
- 39.Zaher H, Fernandez-Salguero PM, Letterio J, et al. The involvement of aryl hydrocarbon receptor in the activation of transforming growth factor-beta and apoptosis. Molecular Pharmacology. 1998;54(2):313-21. [DOI] [PubMed] [Google Scholar]
- 40.Letamendia A, Labbe E, Attisano L. Transcriptional regulation by Smads: crosstalk between the TGF-beta and Wnt pathways. The Journal of Bone and Joint Surgery American Volume. 2001;83-A Suppl 1(Pt 1):S31-9. [PubMed] [Google Scholar]
- 41.Tschumperlin DJ, Liu F, Tager AM. Biomechanical regulation of mesenchymal cell function. Current Opinion in Rheumatology. 2013;25(1):92-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. The Journal of Clinical Investigation. 2007;117(3):524-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito T, Yokota K, Kobayakawa K, et al. Experimental Mouse Model of Lumbar Ligamentum Flavum Hypertrophy. PloS One. 2017;12(1):e0169717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salama R, Sadaie M, Hoare M, et al. Cellular senescence and its effector programs. Genes & Development. 2014;28(2):99-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coppe JP, Desprez PY, Krtolica A, et al. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annual Review of Pathology. 2010;5:99-118. [DOI] [PMC free article] [PubMed] [Google Scholar]


