Abstract
Purpose
Multidrug resistance (MDR) and the subsequent disease relapse are the major causes of childhood acute lymphoblastic leukemia (ALL) related death. The Hedgehog (Hh) signaling pathway can contribute to cancer MDR. In the current study, Smoothened (Smo) was selected as the experimental target due to its importance in the Hh pathway in order to evaluate its probable role in pediatric B-ALL drug resistance.
Patients and methods
The study included 27 pediatric B-ALL and 16 control bone marrow samples. Quantitative RT-PCR was used to investigate the expression levels of Smo and miR-326 as the key players of the Hh pathway. Western blot analysis was performed. The presence of minimal residual disease was studied using PCR-SSCP. The association between Smo expression and drug resistance was analyzed statistically.
Results
Results showed a significant increase in the Smo expression levels in drug-resistant patients in comparison with drug-sensitive children with B-ALL (P=0.0128, AUC=0.82). A considerable negative association between miR-326 and Smo expression levels was identified (r=−0.624, P=0.002). A binomial test confirmed the regulatory role of miR-326 on the translational repression of Smo (P=0.031). Statistics showed no association between Smo and ABCA2 expression levels. However, a significant positive correlation was observed between the Smo and ABCA3 transcripts in the resistant ALL children (r=0.607, P=0.016).
Conclusion
Data revealed the possible oncogenic impact of Smo on leukemogenesis and drug resistance in pediatric B-ALL. Upregulation of Smo was introduced, for the first time, as a prognostic factor for drug resistance in childhood B-ALL. To the best of our knowledge, this is the first study that shows a positive correlation between Smo and ABCA3 expression levels in pediatric B-ALL, explaining a possible mechanism for the development of drug resistance in this cancer. Moreover, the current project revealed a negative modulatory effect of miR-326 on the expression levels of Smo.
Keywords: Hh pathway, Smo, miR-326, ABC transporter
Introduction
Acute lymphoblastic leukemia (ALL) is known as the most prevalent hematological malignancy in children under 15 years of age, where the B-cell lineage subtype is most common.1 Despite recent progress in the development of efficient treatments, disease relapse remains the major cause of childhood cancer-related death.2,3 This relapse is a result of resistance against chemotherapeutic regimens, which is often related to the dysregulation of genes associated with multidrug resistance (MDR).4 One of the most important mechanisms underlying MDR is the increased expression of adenosine triphosphate (ATP)-binding cassette (ABC) transporters which leads to an increase in the efflux of various anti-cancer drugs from cancer cells, resulting in chemotherapy failure.5,6
Hedgehog (Hh) is a conserved developmental signaling pathway with a key regulatory function in embryonic patterning and adult tissue homeostasis, which is of importance in primitive and adult hematopoiesis.7,8 Hh signaling can be initiated by the binding of Hh protein ligands (Sonic, Indian, and Desert) to the cell surface receptor Patched (Ptch).9 This binding releases inhibitors of another transmembrane protein called Smoothened (Smo), a signal transducer, which eventually results in the activation and nuclear translocation of Gli transcription factors (Gli 1, Gli 2, and Gli 3) at target promoters.7,9 This process transactivates Hh responsive genes, including Gli 1, Ptch 1, Cyclin D, Bcl-2, and some other genes associated with stem cell differentiation, such as Sox 2, Oct 4, Bmi 1, and Nanog.10,11,12 Aberrant activation of the pathway is observed in a wide range of human cancers, including glioblastoma, basal cell carcinoma (BCC), chronic myeloid leukemia (CML), and multiple myeloma (MM).13–15 It has been described that dysregulation of Hh signaling in some cancers may be due to the overexpression of Smo, the key signal transducer involved in this pathway, which can result in Gli activation and subsequent contributions to various aspects of oncogenesis such as maintenance of cancer stem cells (CSCs) and drug resistance.11,16
MicroRNAs (miRNAs) are a large family of single-stranded non-coding small RNAs in their mature form.17,18 They can interact with their complementary target sequence within the 3ʹ untranslated region (3ʹ-UTR) of mRNA and regulate gene expression by translational repression or degradation of the target mRNA.19,20 MiRNAs play key roles in modulating various fundamental, physiological, and biological processes in normal cells.21,22 However, dysregulation of these tiny, small transcripts may contribute to cancer.20,21 In our previous study, miR-326 was introduced as a tumor-suppressor gene which is downregulated in children with ALL and ALL drug resistance.23
As the Hh signaling pathway can lead to the maintenance of stemness signatures through targeting pluripotency and drug resistance-related genes, we selected Smo as our experimental target because of its importance in this pathway. To the best of our knowledge, this is the first study through which we verified the upregulation of the signal transducer Smo in patients with B-ALL and those who were resistant to chemotherapy. Subsequently, we confirmed the significant negative correlation between the increased expression levels of Smo and decreased concentrations of miR-326. Furthermore, we unveiled the probable regulatory role of Smo on ABCA3 expression as a probable target of the Hh pathway and involved gene in drug resistance.
Materials and methods
Patients and control samples
The study included 27 pediatric B-ALL and 16 non-cancer control bone marrow samples, obtained from children referred to Sayed-ol-Shohada Hospital (Isfahan, Iran) during a two-year period, from 2014 to 2015. The study was conducted in accordance with the Declaration of Helsinki, and permitted by the Ethics Committee of the University of Isfahan (agreement number IR.UI.REC.1397.151). Informed written consents were obtained from the children’s parents prior to participation in the study. B-cell acute lymphoblastic leukemia was diagnosed clinically and confirmed pathologically. Nineteen patients were newly diagnosed and 8 patients were administered to the hospital during the period of the study and diagnosed with ALL relapse. Controls were individuals with no evidence of cancer. About 2–5 mL of heparinized bone marrow samples were collected from B-ALL patients and controls for further analysis. Mononuclear cell separation was performed by sedimentation on lymphoprep density gradients (Axis Shailed Diagnostics Ltd., Oslo, Norway), according to the manufacturer recommended protocol.
RNA extraction and cDNA synthesis
In accordance with the manufacturer’s instructions, total RNA was extracted using TRizol reagent (Ambion, Carlsbad, CA) and treated with RNase-free DNase (Fermentas, Vilnius, Lithuania). Then, 2 μg of total RNA was used for cDNA synthesis, utilizing the PrimeScriptTM RT reagent kit (Takara, Shiga, Japan) with random hexamers and oligo dT primers. The reverse transcription reaction was carried out at 37°C for 15 mins and followed by reverse transcriptase enzyme inactivation at 85°C for 5 s. The obtained cDNA was stored at −20°C for further analysis.
Real-time PCR analysis
To quantify gene expression, qRT-PCR reactions were performed using ExiLENT SYBR Green master mix using Chromo4TM system (Bio-Rad, Foster City, California), as per the manufacturer’s instructions. The expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression as an endogenous control. Thermal cycling was performed using the following cycling conditions: 95°C pre-denaturation for 5 mins followed by 95°C denaturation for 20 s, 60°C annealing for 30 s, and 72°C extension for 30 s. The samples were run in triplicates. The relative expression was measured and reported using the 2−ΔΔCt method. To confirm the presence of the Smo primers, blasting pre-used primers in bone marrow and UCSC database were exploited and then primers were double-checked with gel electrophoresis. The sequences of the primers are shown in Table 1.
Table 1.
Primer sequences of Smo and GAPDH
| Gene | Primer Sequence (5ʹ to 3ʹ) | Primer length | Amplicon length | Tm | GC% |
|---|---|---|---|---|---|
| Smo | F: TTACCTTCAGCTGCCACTTCTAC | 23 bp | 321 bp | 60.31 | 47.83 |
| R: CCTTGGCAATCATCTTGCTCTTC | 23 bp | 60.18 | 47.83 | ||
| GAPDH | F: CAAGGCTGAGAACGGGAA | 18 bp | 90 bp | 56.88 | 55.56 |
| R: GCATCGCCCCACTTGATTTT | 20 bp | 59.47 | 50 |
Response to chemotherapy
Newly diagnosed B-ALL patients were treated based on the Australian and New Zealand Children’s Cancer Study Group ALL study protocol 8 (http://www.anzctr.org.au/trial_view.aspx?ID=1568). For detection of the treatment response in these patients, the minimal residual disease (MRD) was determined using PCR-SSCP (single-strand conformation polymorphism) analysis for immunoglobulin heavy chain (IgH) and T-cell receptor gamma (TcRγ) gene rearrangements undertaken exactly one year after treatment. Observation of monoclonality, referred to as MRD+, represented drug resistance and non-response chemo treatment. Drug-sensitive patients were nominated as MRD− individuals.
Western blot analysis
To investigate the potential role of the Smo protein on drug resistance, patients were divided into two groups of resistant (MDR) and sensitive (MRD-), and Western blot analysis was performed on the derived samples. Total protein was extracted from cells using a lysis buffer. Bradford protein assay was performed as previously described,4 and samples with enough concentrations of protein (11 MDR and 7 MRD-) were subjected to electrophoresis on 10% SDS-PAGE, and then transferred from the gel to a polyvinylidene difluoride (PVDF) membrane. Non-specific sites on the membrane were blocked with 10% skim milk at room temperature for 1 hr. The membrane was then incubated with primary antibody for Smo (1:200 Santa Cruz Biotechnology, CA) at 4°C overnight. B-Actin monoclonal antibody (Santa Cruz Biotechnology, CA) was used as the loading control. After three washes, the membrane was incubated with secondary anti-mouse IgG HRP (1:1,000, Santa Cruz Biotechnology, CA) at room temperature for 1 hr. Finally, proteins were detected on sensitive radiology films in a darkroom using ECL solution (Amersham, Piscataway, New Jersey).
Statistical analysis
All statistical analyses were performed using SPSS23.0 and GraphPad Prism7 softwares. The Kolmogorov–Smirnov normality test (KS test) and Shapiro–Wilk test were applied to evaluate the normality of data distribution. Comparisons of the data from each group with the control were carried out to assess the statistical significance of the difference by Kruskal–Wallis, Mann–Whitney, and Binomial nonparametric test. The receiver operating characteristic (ROC) curves and the area under the ROC (AUC) were depicted to evaluate the specificity and sensitivity of Smo as a diagnostic and/or prognostic biomarker for B-ALL. The correlation between parameters was analyzed through a Spearman’s coefficient calculation. All data were expressed as mean±standard error of mean (SEM). P-values equal to or less than 0.05 were considered to be statistically significant.
Results
Nineteen bone marrow samples from de novo non-Philadelphia-positive B-ALL children <12 years of age, comprising 8 MRD+ and 11 MRD- patients, were entered into the study and compared with 16 bone marrow samples from individuals (including 9 males and 7 females) with no evidence of malignancy. The mean age of patients and controls was 5.3±0.117 and 4.8±0.9 years, respectively (mean±SEM). No significant dissimilarity was observed in the distribution of age and sex between cases and controls (P>0.05). Eight relapsed patients and 8 MRD+ patients who exhibited MDR after receiving chemotherapy were categorized as the MDR patient group. Patients’ clinical information including sex and white blood cell (WBC) count is presented in Table 2.
Table 2.
Primary data of the patients included in the study
| Patientcharacteristics | Number/total patients | Percentage | |
|---|---|---|---|
| Sex | Male | 13/27 | 48 |
| Female | 14/27 | 52 | |
| MRD | MRD+ | 8/27 | 29.63 |
| MRD- | 11/27 | 40.74 | |
| Relapse | in Bone marrow | 8/27 | 29.63 |
| WBC | >50,000/µL whole blood | 10/27 | 37 |
Note: MRD, minimal residual disease determined in new case patients one year after treatment.
Abbreviations: MRD, minimal residual disease; WBC, white blood cell.
Diagnostic significance of Smo in pediatric B-ALL and immunoblotting analysis
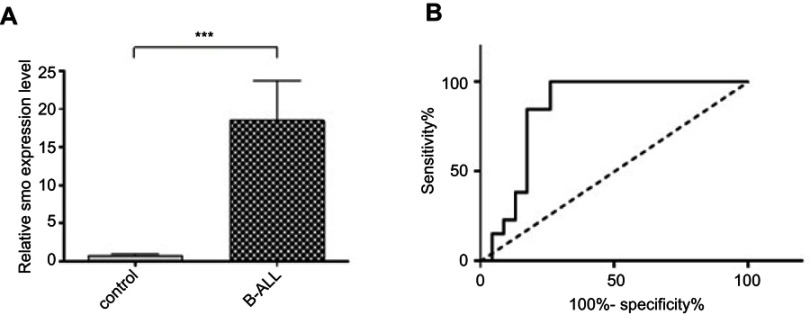
To evaluate the prognostic value of Smo as a signal transducer of the Hh pathway, we screened its expression in patients with B-ALL. The comparison between transcript levels of Smo in patient and control groups revealed that Smo was highly expressed in pediatric B-ALL, with a P-value of 0.0002 (Figure 1A). Then, ROC curve analysis was performed to evaluate the suitability of this oncogene for discriminating between ALL individuals and non-cancer controls. The total area of 0.84 under the curve (AUC) proved the effectiveness and accuracy of this measurement (Figure 1B). It demonstrated that the Smo expression level could serve as a biomarker to distinguish between B-ALL patients and controls (P=0.0007).
Figure 1.
Smo expression level in new case and control groups. (A) Significant upregulation of Smo expression relative to GAPDH as a housekeeping gene in B-ALL subjects compared with the control group using RT-PCR assay (***P=0.0002). (B) Diagnostic potential of Smo. ROC curve analyses showed that the mRNA Smo expression levels could distinguish B-ALL children from non-leukemic controls (AUC=0.84, P=0.0007).
Abbreviations: B-ALL, B-cell acute lymphoblastic leukemia; ROC, receiver operating characteristic.
Prognostic significance of Smo expression in pediatric B-ALL
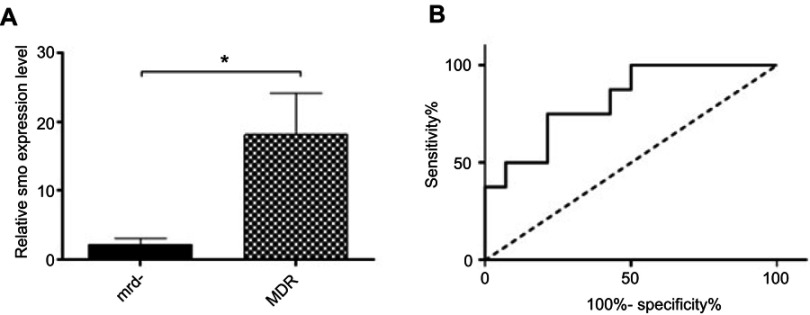
To determine the prognostic value of Smo and its possible effect on drug resistance in pediatric B-ALL, we compared the transcript levels of this oncogene in drug-sensitive (MRD-) patients and drug-resistant or MDR group (MRD+ and relapsed patients). Results demonstrated a significant increase in Smo expression levels in the MDR group compared with the MRD- group (P=0.0128) (Figure 2A). Using ROC curve analysis, the AUC of 0.82 introduced the cellular Smo levels as a negative prognostic biomarker with a predictive ability to discriminate the MDR group from the MRD- patients (P=0.0117) (Figure 2B).
Figure 2.
Comparison between the MDR (MRD+ and relapsed) and MRD- groups with respect to the Smo expression levels. (A) Significant increased Smo expression in MDR patients, known as resistant group, compared with MRD-, considered as sensitive group (*P=0.0128). (B) Prognostic potential of Smo using ROC curve analyses. Smo transcript level could significantly discriminate MDR patients from MRD- ones (AUC of 0.82, P=0.0117).
Abbreviations: MDR, multidrug resistance; ROC, receiver operating characteristic.
Western blotting to investigate the correlation between MDR and the protein expression levels of Smo
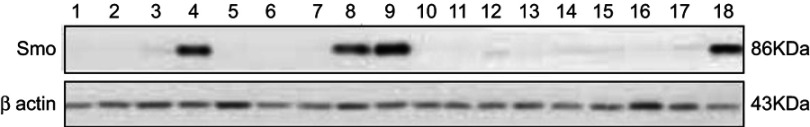
Western blot analysis was performed on the available samples of two groups of drug-resistant and drug-sensitive ALL patients. Results showed that only 4 out of 13 (30.7%) patients with high transcript levels of Smo had a significant expression of the Smo protein (Figure 3). Interestingly, all those samples belonged to the leukemic patients with MDR to chemotherapy (Table 3).
Figure 3.
Western blot analysis of Smo protein expression in 18 available samples for protein analysis. Numbers 1–9, 17, and 18=Samples of patients with multidrug resistance (MDR), Numbers 10–16=Samples of sensitive patients to chemotherapy (MRD-).
Table 3.
The expression profile of Smo at the mRNA and protein levels in 18 patients with B-ALL
| Smo mRNA+ | Smo protein+ | miR-326- | Total | |
|---|---|---|---|---|
| MDR | 10 | 4 | 7 | 11 |
| MRD- | 3 | 0 | 5 | 7 |
| Total | 13 | 4 | 12 | 18 |
Note: N, number of samples, Smo mRNA+, samples with overexpression of Smo at the mRNA levels; Smo protein+, samples with overexpression of Smo at protein levels; miR-326-, samples with downregulation of miR-326.
Abbreviations: MDR, multidrug resistance; B-ALL, B-cell acute lymphocytic leukemia.
In the next step, to examine the possible role of miR-326 in regulating the Smo transcripts, resulting in the observed adverse gene protein levels, a non-parametric binomial test was performed. Samples were divided into two groups: a group with an inverse correlation between miR-326 and Smo mRNA expression levels (Table 4, group 1) and a group with a direct correlation between the miRNA and Smo transcript levels (Table 4, group 2). The P-value of 0.031 suggests that samples distribution between the abovementioned groups is significantly different so that the first group with the higher frequency of 78% demonstrates the characteristics of the statistical study population.
Table 4.
Non-parametric binomial test to examine the correlation between miR-326 and Smo expression levels
| Groups | Number | Observed proportion | P-value |
|---|---|---|---|
| Group 1 | 14 | 0.78 | 0.031 |
| Group 2 | 4 | 0.22 | |
| Total | 18 | 1.00 |
Note: Group 1, patients with inverse correlation between miR-326 and Smo expression levels; Group 2, patients with direct correlation between miR-326 and Smo expression levels.
Downregulation of miR-326 and upregulation of Smo in drug-resistant pediatric B-ALL
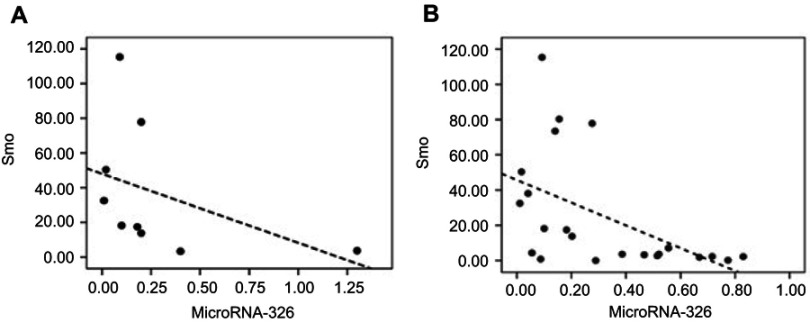
In accordance with our previous study, significant downregulation of miR-326 was confirmed in ALL patients compared with the non-leukemic controls.23 To evaluate the correlation between miR-326 and the Hh signaling pathway in resistant childhood ALL, statistical analysis was performed with consideration to the expression levels of miR-326 and Smo as the signal transducer. As shown in Figure 4A, the expression levels of miR-326 were inversely correlated with the expression levels of Smo in leukemic patients (r=−0.68, P=0.045) (Figure 4A). Our data suggest that the upregulation of Hh signaling in resistant B-ALL patients could be due to the decreased expression of miR-326, and consequently, upregulation of Smo.
Figure 4.
Possible correlation between the mRNA expression levels of Smo and miR-326 was determined using a Spearman’s correlation test. (A) Inverse correlation between the expression levels of Smo and miR-326 in the multidrug resistant, MDR, ALL patients (r=−0.68, P=0.045). (B) Inverse correlation between the mRNA expression levels of Smo and miR-326 in patients with B-ALL (r=−0.626, P=0.002).
Abbreviations: MDR, multidrug resistance; B-ALL, B-cell acute lymphocytic leukemia.
Statistical analyses were also carried out for all the ALL children, regardless of their response to treatment. Interestingly, results showed a similar significance (r=−0.624, P=0.002) (Figure 4B).
Increased mRNA expression levels of ABCA3 were positively correlated with the expression levels of Smo in B-ALL children resistant to chemotherapy
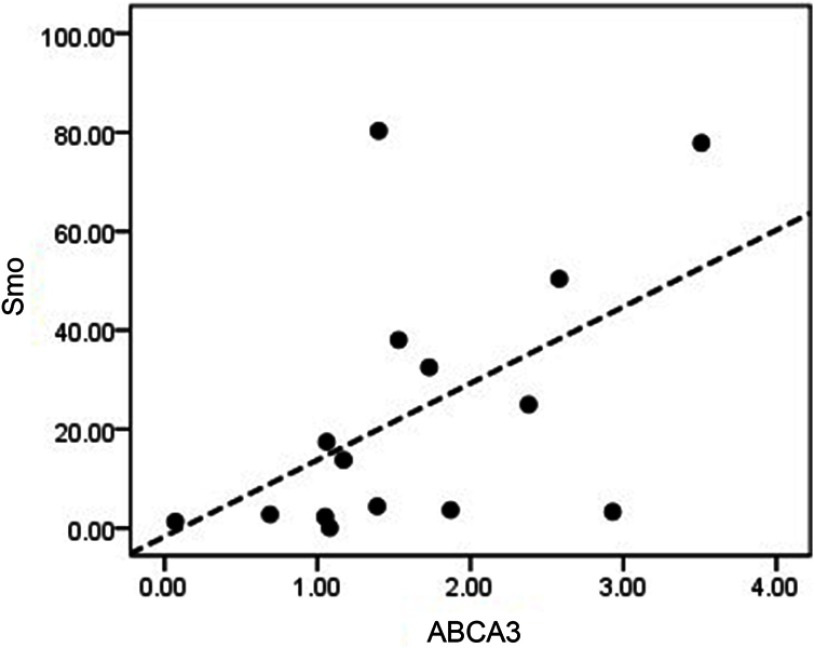
A recent study by our group has determined the role of ABCA2 and ABCA3 overexpression as a poor prognostic biomarker in multidrug-resistant childhood ALL.23 To investigate whether the Hh signaling pathway could exert its impact on childhood B-ALL drug resistance through regulating the expression of the aforementioned ABC transporters, we statistically analyzed the correlation between the mRNA expression levels of Smo and the drug efflux pumps ABCA2 and ABCA3 in drug-resistant B-ALL children. Based on a Spearman’s correlation test, the expression levels of Smo and ABCA2 showed no significant association. However, a positive correlation was revealed between the expression levels of Smo and the ABCA3 transporter. (r=0.607, P=0.016) (Figure 5).
Figure 5.
Investigating any possible association between the mRNA expression levels of Smo and ABCA3 in drug-resistant patients. Spearman’s correlation test showed a direct correlation between the mRNA expression levels of Smo and ABCA3 genes (r=0.607, P=0.016).
Discussion
Despite recent advances in the treatment of childhood ALL, 20% of the patients still present with treatment failure and relapse.24 For a majority of children who encounter relapse, achieving a complete remission is impossible.3 The current study introduces a novel biomarker and a regulatory microRNA which can be implicated in new cancer therapeutic strategies.
The Hh signaling pathway is likely to play a crucial role in hematopoietic system regulation, as the stimulation of the Hh ligands is necessary for hematopoiesis in the animal models with Hh deficiency.8,25,26 Furthermore, activation of the Hedgehog pathway has been observed in different neoplasms, suggesting that it plays an important role in self-renewal regulation of tumor cells.10,11 In the current study, considering the functional role of Smo in activating the Hh pathway, we focused on the expression of this gene. Moreover, overexpression of Smo could result in the induction of the self-renewal properties of leukemic CSCs and cause subsequent drug resistance.11,27 In addition, the inhibition of Smo could inhibit proliferation and invasion of cancer cells and enhance sensitivity to chemotherapy.28,29 Results of this study revealed, for the first time, a significant overexpression of Smo at the onset of disease, indicating its potential role as an oncogene in pediatric B-ALL (de novo patients in comparison with control group, P=0.0002). In addition, the area under the ROC curve (0.84) indicated a high specificity and sensitivity of Smo expression levels for discriminating between children with B-ALL and non-cancer individuals (P=0.0007). On the other hand, our findings showed a significant increase in the mRNA expression levels of Smo in drug-resistant patients in comparison with drug-sensitive children with B-ALL (P=0.0128). We also evaluated the specificity and sensitivity of Smo’s impact on drug resistance using ROC curve analysis, which yielded an AUC of 0.82 in discriminating the drug-resistant group (including MRD+ and relapsed patients) from the MRD- ALLs (P=0.0117). The larger total area under the curve (AUC) value indicated a better overall performance of the medical test in correctly distinguishing between the two situations of resistance or sensitivity to chemotherapy drugs. Accordingly, it could be hypothesized that Smo might have a prognostic role in children with B-ALL.
In the next step, to investigate the correlation between drug resistance and Smo expression at protein levels, samples were divided into two different categories of drug-resistant and drug-sensitive patients. Subsequently, Western blotting was performed on the samples with enough number of cells pertaining to the two aforementioned groups. It is worth mentioning that all samples without the overexpression of Smo at protein levels, regardless of their mRNA expression levels, belonged to the MRD- patients. Furthermore, a total number of 4 patients showed upregulation of Smo at both mRNA and protein levels. Interestingly, all these patients were multidrug resistant to the ALL treatment and belonged to the MDR group. These data indicated the possible role of Smo in poor response to treatment in B-ALL. Regarding the patients with Smo overexpression at mRNA but not protein levels, the possible impact of miR-326 on the translational repression of Smo was investigated using a non-parametric binomial test. The rationale in selecting miR-326 in this context was based on the results of our previous study demonstrating the role of miR-326 in leukemogenesis as a tumor-suppressor, as well as its negative impact on MDR in children with ALL.23 As shown in Table 4, the P-value of 0.031 suggested that among the 18 available cases for Western blotting, samples distribution between the first group (including patients with inverse correlation between miR-326 and Smo mRNA expression levels and the second group containing patients with direct correlation of the aforementioned genes) was significantly different. Therefore, group one with the highest frequency (78%) demonstrated the characteristic of the statistical study population. These results could be considered as confirming evidence for the regulatory role of miR-326 on MDR through the modulation of Smo, which is consistent with the results of Tang et al’s study on the multiple myeloma patients.29 However, negative response to chemotherapy in MDR patients with a lack of Smo proteins may have been caused by the interference of other molecular mechanisms contributing to drug resistance.30
Despite the fact that there have already been several studies regarding miR-326 and its functional importance in the Hh pathway, little is known about its interaction with this pathway in childhood B-ALL and MDR. A recent study demonstrated that miR-326 could target the Smo oncogene to inhibit the biological behaviors and stemness in glioma cells.31 Others have found that it suppresses the activity of the Hh pathway, which in turn may influence drug resistance.32 In the present study, for the first time, we emphasized the role of miR-326 in the MDR of B-ALL children through regulating the expression of Smo, the key player in the Hh signaling pathway. This assertion was supported by a Spearman’s correlation coefficient data analysis demonstrating a significant inverse correlation between miR-326 and Smo expression levels (r=−0.624, P=0.002). These findings are in line with the results of another study by Yin et al, in which Smo was verified as a direct target of miR-326 through bioinformatics and functional experiments.32 According to our data, miR-326 can be suggested as a potential candidate for targeted therapy, contributing to improved treatment outcome in drug-resistant B-ALL children. The pertinent point to note is that miR-326, like other miRNAs, may target a number of genes. Whether these other targets are involved in the regulation of drug resistance in B-ALL is a question that requires further delineation.
Several studies have described the probable mechanisms involved in drug resistance, in which the Hh signaling pathway plays an indispensable role. Furthermore, this pathway can contribute to MDR via different ways.16,33,34 According to an in vivo study performed on gastric cancer, the overexpression of ABCG2 can cause drug resistance in mouse models as a direct target of the Hh signaling pathway.35 However, our previous study on this ABC transporter showed no significant upregulation in ALL patients.36 On the other hand, taking into account the role of the Hedgehog pathway in the regulation of the stemness-related genes, including Oct 4, Sox 2, Bmi 1, and Nanog, oncogenic activation of the Hh pathway in patients can undoubtedly lead to cancer stem cell survival.10,37,38 As a consequence, considering the great ability of these cells to express drug resistance-related genes, such as ABC transporters, these cells can remain even after chemotherapy.39,40 Excitingly, our previous study exhibited a positive correlation between ABCA2/ABCA3 transporters and drug resistance in childhood ALL.4 Consequently, a statistical analysis was carried out in order to assess any possible association between the mRNA expression profiles of Smo and the abovementioned drug pumps. Data revealed no impact on ABCA2, while a significant positive correlation was observed between the Smo and ABCA3 transcripts in the resistant ALL children (r=0.607, P=0.016). A schematically possible regulatory axis is suggested to exist in pediatric ALL leukemic cells among miR-326, Smo and the ABCA3 transporter, which may contribute to drug resistance (Figure 6). Moreover, the correlation between miR-326 and ABCA2, that was previously shown by our group (r=−0.458 and P=0.01), is also demonstrated to complete this proposed contribution.5 Further experiments are required to validate the direct interactions between the indicated molecules.
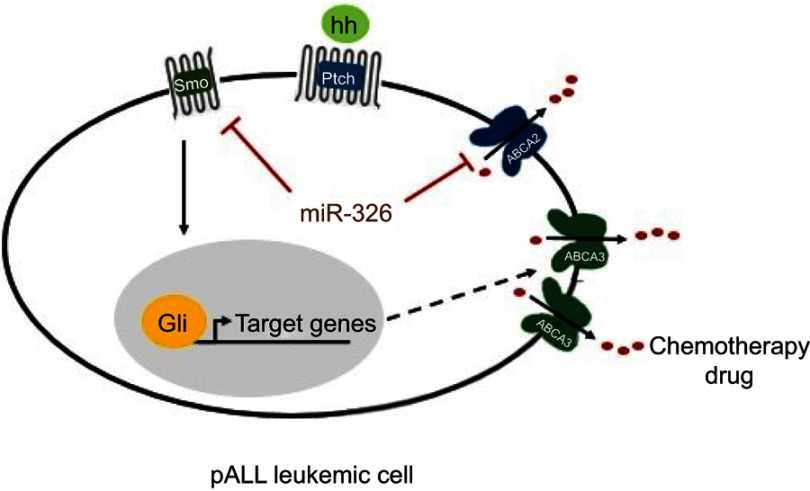
Figure 6.
Possible mechanism of drug resistance related to miR-326 in pediatric acute lymphoblastic leukemia (pALL) leukemic cells. Smo gene targeted by miR-326 may regulate the stemness-related target genes of Hh pathway. It may contribute to leukemic cells survival and, subsequently, overexpression of the drug resistance gene, ABCA3, followed by efflux of the prescribed chemotherapy drugs. In addition, it is likely that miR-326 completes its role in drug resistance through the regulation of ABCA2 transporter. Black arrow, stimulation; red blunt-ended arrow, direct inhibition; dashed arrow, unconfirmed indirect effect; hh, protein ligand; pALL, pediatric acute lymphoblastic leukemia.
In brief, this study provides evidence, for the first time, showing the possible oncogenic impact of Smo, as a key player of the hedgehog pathway, on leukemogenesis and drug resistance in pediatric B-ALL. Furthermore, a Spearman’s correlation test revealed a positive correlation between the high expression levels of Smo and the ABCA3 transporter, suggesting a regulatory role for the Hh signaling pathway in resistance to chemotherapy drugs in this cancer. Eventually, a negative modulatory effect of miR-326 on the expression levels of Smo introduced this microRNA as a potential novel target for the treatment of childhood B-ALL. Further delineation of the links between miR-326, Smo, and ABCA3 should provide novel approaches to new diagnostics and therapeutics.
Conclusion
In pediatric ALL, further reinforcement of the current chemotherapy regimens is unlikely to improve cure rate, due to the accompanying increased toxicity. Therefore, novel anti-ALL agents are needed to overcome chemotherapy resistance and reduce non-specific toxicity. Importantly, in this study we elucidated, for the first time, that upregulation of Smo was involved in resistance to the chemotherapeutic drugs in childhood B-ALL. A positive correlation between Smo and ABCA3 expression levels was also found in pediatric B-ALL samples, explaining a possible mechanism for the development of drug resistance in this cancer. This study provides new evidence about the role of Smo in pediatric B-ALL and development of MDR and suggests a potential relationship between the Hh signaling pathway and leukemia relapse.
Acknowledgments
Authors are indebted to professor Alireza Moafi for kindly providing the samples, as well as the children and parents who participated in the study, without whom this research would not be possible.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Rytting ME, Jabbour EJ, O’brien SM, Kantarjian HM. Acute lymphoblastic leukemia in adolescents and young adults. Cancer. 2017;123(13):2398–2403. doi: 10.1002/cncr.30624 [DOI] [PubMed] [Google Scholar]
- 2.Wu C, Li W. Genomics and pharmacogenomics of pediatric acute lymphoblastic leukemia. Crit Rev Oncol Hematol. 2018;126:100–111. doi: 10.1016/j.critrevonc.2018.04.002 [DOI] [PubMed] [Google Scholar]
- 3.Tzoneva G, Dieck CL, Oshima K, et al. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature. 2018;553(7689):511. doi: 10.1038/nature25186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aberuyi N, Rahgozar S, Dehaghi ZK, Moafi A, Masotti A, Paolini A. The translational expression of ABCA2 and ABCA3 is a strong prognostic biomarker for multidrug resistance in pediatric acute lymphoblastic leukemia. Onco Targets Ther. 2017;10:3373. doi: 10.2147/OTT.S140488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghodousi ES, Rahgozar S. MicroRNA-326 and microRNA-200c: two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J Cell Biochem. 2018;119:1–9. [DOI] [PubMed] [Google Scholar]
- 6.Jaramillo AC, Al Saig F, Cloos J, Jansen G, Peters GJ. How to overcome ATP-binding cassette drug efflux transporter-mediated drug resistance. Cancer Drug Resist. 2018;1:6–29. doi: 10.20517/cdr.2018.02 [DOI] [Google Scholar]
- 7.Fattahi S, Langroudi MP, Akhavan‐Niaki H. Hedgehog signaling pathway: epigenetic regulation and role in disease and cancer development. J Cell Physiol. 2018;233(8):5726–5735. doi: 10.1002/jcp.26506 [DOI] [PubMed] [Google Scholar]
- 8.Trowbridge JJ, Scott MP, Bhatia M. Hedgehog modulates cell cycle regulators in stem cells to control hematopoietic regeneration. Proc Natl Acad Sci. 2006;103(38):14134–14139. doi: 10.1073/pnas.0604568103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu F, Zhang Y, Sun B, McMahon AP, Wang Y. Hedgehog signaling: from basic biology to cancer therapy. Cell Chem Biol. 2017;24(3):252–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clement V, Sanchez P, De Tribolet N, Radovanovic I, Altaba AR. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Current Biol. 2007;17(2):165–172. doi: 10.1016/j.cub.2006.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochrane C, Szczepny A, Watkins D, Cain J. Hedgehog signaling in the maintenance of cancer stem cells. Cancers. 2015;7(3):1554–1585. doi: 10.3390/cancers7030851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The role of the Hedgehog signaling pathway in cancer: a comprehensive review. Bosn J Basic Med Sci. 2018;18(1):8. doi: 10.17305/bjbms.2018.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacouture ME, Dréno B, Ascierto PA, et al. Characterization and management of hedgehog pathway inhibitor-related adverse events in patients with advanced basal cell carcinoma. Oncologist. 2016;21(10):1218–1229. doi: 10.1634/theoncologist.2016-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babashah S, Sadeghizadeh M, Hajifathali A, et al. Targeting of the signal transducer Smo links microRNA‐326 to the oncogenic Hedgehog pathway in CD34+ CML stem/progenitor cells. Int J Cancer. 2013;133(3):579–589. doi: 10.1002/ijc.28043 [DOI] [PubMed] [Google Scholar]
- 15.Blotta S, Jakubikova J, Calimeri T, et al. Canonical and noncanonical Hedgehog pathway in the pathogenesis of multiple myeloma. Blood. 2012;120(25):5002–5013. doi: 10.1182/blood-2011-07-368142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sims-Mourtada J, Izzo J, Ajani J, Chao K. Sonic Hedgehog promotes multiple drug resistance by regulation of drug transport. Oncogene. 2007;26(38):5674. doi: 10.1038/sj.onc.1210356 [DOI] [PubMed] [Google Scholar]
- 17.Gounaris-Shannon S, Chevassut T. The role of miRNA in haematological malignancy. Bone Marrow Res. 2013;2013:1–12. doi: 10.1155/2013/269107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pouyanrad S, Rahgozar S, Ghodousi ES. Dysregulation of miR-335-3p, targeted by NEAT1 and MALAT1 long non-coding RNAs, is associated with poor prognosis in childhood acute lymphoblastic leukemia. Gene. 2019;692:35–43. doi: 10.1016/j.gene.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 19.Shah N, Bowles K, Rushworth SA, MacEwan D. Understanding the role for miRNA in human leukemia. RNA Dis. 2015;2(1). [Google Scholar]
- 20.Wang Y, He J, Wang Y, Yu H. MicroRNA-138 suppresses cell proliferation, migration and invasion by targeting smoothened (SMO) in hepatocellular carcinoma. Int J Clin Exp Med. 2017;10(9):13281–13289. [Google Scholar]
- 21.Xu Z, Huang C, Hao D. MicroRNA-1271 inhibits proliferation and promotes apoptosis of multiple myeloma cells through inhibiting smoothened-mediated Hedgehog signaling pathway. Oncol Rep. 2017;37(2):1261–1269. doi: 10.3892/or.2016.5304 [DOI] [PubMed] [Google Scholar]
- 22.Ultimo S, Martelli AM, Zauli G, Vitale M, Calin GA, Neri LM. Roles and clinical implications of microRNAs in acute lymphoblastic leukemia. J Cell Physiol. 2018;233(8):5642–5654. doi: 10.1002/jcp.26290 [DOI] [PubMed] [Google Scholar]
- 23.Ghodousi ES, Rahgozar S. MicroRNA‐326 and microRNA‐200c: two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J Cell Biochem. 2018;119(7):6024–6032. doi: 10.1002/jcb.26800 [DOI] [PubMed] [Google Scholar]
- 24.Ma X, Edmonson M, Yergeau D, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604. doi: 10.1038/ncomms7604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao C, Chen A, Jamieson CH, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458(7239):776. doi: 10.1038/nature07737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dierks C, Beigi R, Guo G-R, et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14(3):238–249. doi: 10.1016/j.ccr.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 27.Lin TL, Wang QH, Brown P, et al. Self-renewal of acute lymphocytic leukemia cells is limited by the Hedgehog pathway inhibitors cyclopamine and IPI-926. PLoS One. 2010;5(12):e15262. doi: 10.1371/journal.pone.0015262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, Lv Y, Wang L, et al. Inhibition of hedgehog pathway reveals the regulatory role of SMO in gastric cancer cells. Tumor Biol. 2017;39(7):1010428317715546. doi: 10.1177/1010428317715546 [DOI] [PubMed] [Google Scholar]
- 29.Tang B, Xu A, Xu J, et al. MicroRNA‐324‐5p regulates stemness, pathogenesis and sensitivity to bortezomib in multiple myeloma cells by targeting hedgehog signaling. Int J Cancer. 2018;142(1):109–120. doi: 10.1002/ijc.31041 [DOI] [PubMed] [Google Scholar]
- 30.Ramos P, Bentires-Alj M. Mechanism-based cancer therapy: resistance to therapy, therapy for resistance. Oncogene. 2015;34(28):3617. doi: 10.1038/onc.2014.462 [DOI] [PubMed] [Google Scholar]
- 31.Du W, Liu X, Chen L, et al. Targeting the SMO oncogene by miR-326 inhibits glioma biological behaviors and stemness. Neuro-oncology. 2014;17(2):243–253. doi: 10.1093/neuonc/nou217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin S, Du W, Wang F, et al. MicroRNA-326 sensitizes human glioblastoma cells to curcumin via the SHH/GLI1 signaling pathway. Cancer Biol Ther. 2018;19(4):260–270. doi: 10.1080/15384047.2016.1250981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511(7507):90. doi: 10.1038/nature13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu M, Gong A, Yang H, et al. Sonic hedgehog-glioma associated oncogene homolog 1 signaling enhances drug resistance in CD44+/Musashi-1+ gastric cancer stem cells. Cancer Lett. 2015;369(1):124–133. doi: 10.1016/j.canlet.2015.08.005 [DOI] [PubMed] [Google Scholar]
- 35.Singh RR, Kunkalla K, Qu C, et al. ABCG2 is a direct transcriptional target of hedgehog signaling and involved in stroma-induced drug tolerance in diffuse large B-cell lymphoma. Oncogene. 2011;30(49):4874. doi: 10.1038/onc.2011.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahgozar S, Moafi A, Abedi M, et al. mRNA expression profile of multidrug-resistant genes in acute lymphoblastic leukemia of children, a prognostic value for ABCA3 and ABCA2. Cancer Biol Ther. 2014;15(1):35–41. doi: 10.4161/cbt.26603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batsaikhan B-E, Yoshikawa K, Kurita N, et al. Cyclopamine decreased the expression of sonic hedgehog and its downstream genes in colon cancer stem cells. Anticancer Res. 2014;34(11):6339–6344. [PubMed] [Google Scholar]
- 38.Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, Fields AP. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell. 2014;25(2):139–151. doi: 10.1016/j.ccr.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15(4):338. doi: 10.1038/ncb2717 [DOI] [PubMed] [Google Scholar]
- 40.Takebe N, Warren RQ, Ivy SP. Breast cancer growth and metastasis: interplay between cancer stem cells, embryonic signaling pathways and epithelial-to-mesenchymal transition. Breast Cancer Res. 2011;13(3):211. doi: 10.1186/bcr3052 [DOI] [PMC free article] [PubMed] [Google Scholar]