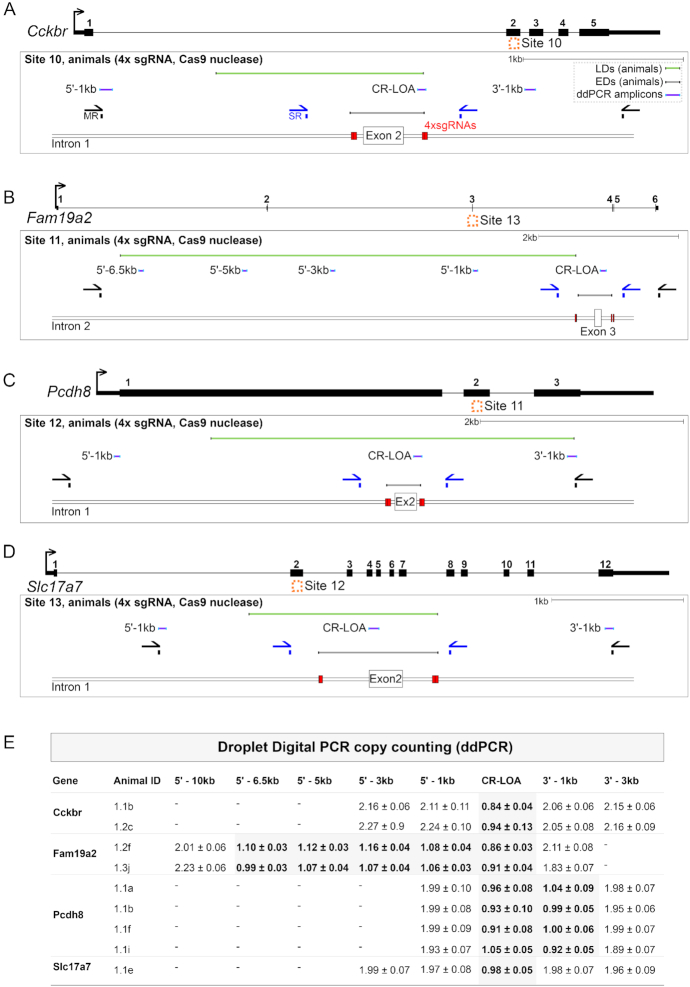
Figure 4.
Larger deletions when genome editing in mouse embryos. (A–D) Locus maps of CRISPR/Cas9 strategies to delete Sites 10–13, corresponding to the genes Cckbr, Fam19a2, Pcdh8 and Slc17a7 respectively. Schematics show the positions of S-R PCR primers (SR, blue), M-R PCR primers (MR, black), sgRNAs (red boxes), ddPCR amplicons (purple lines) and LDs (green lines). (E) Copy counting results from ddPCR experiments. Assays against the wild type genomic sequence were designed in the critical region (CR-LOA) and at 1–3 kb intervals in the 5′ or 3′ direction distal to sgRNA cut sites (e.g. a 5′-1 kb amplicon is located 1 kb in the 5′ direction of the sgRNA cut sites). Each row corresponds to an animal where no deletion was detected by S-R PCR.