Fig. 1.
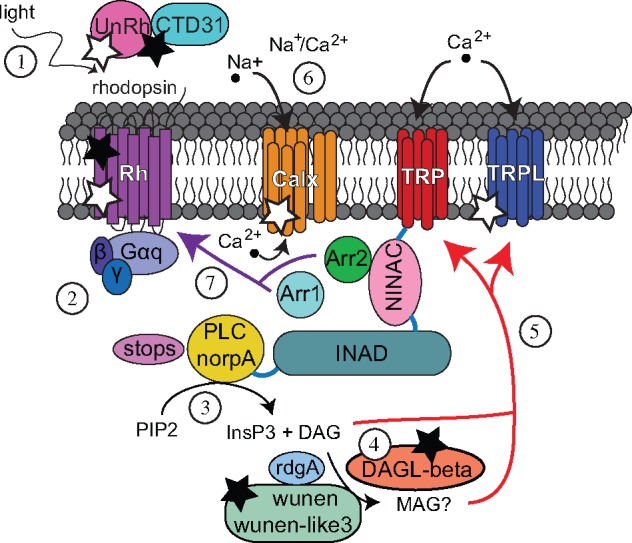
—Speculative model of the lepidopteran phototransduction cascade. Most lepidopteran vision genes are single-copy orthologs of D. melanogaster genes except in chromophore binding, photoisomerization, and diacylglycerol metabolism. 1) Light activates rhodopsin by a conformational change of the chromophore from 11-cis 3-hydroxyretinal to all-trans. The chromophore is transported by Hme CTD31 and photoisomerized from all-trans 3-hydroxyretinal to 11-cis by the unclassified opsin (UnRh), orthologs of which are not found in D. melanogaster. 2) Gαq is released from a G-protein complex of three subunits (α, β, and γ) and activates phospholipase C (PLC). 3) PLC hydrolyzes PIP2 to produce inositol 1,4,5-trisphosphate (InsP3) and diacylglycerol (DAG). 4) Diacylglycerol lipase (DAGLβ) hydrolyzes DAG to produce MAG. In D. melanogaster, DAGLα (inaE) hydrolyzes DAG. DAG levels are also regulated by rdgA and, perhaps, wunen or wunen-like 3. In D. melanogaser, lazaro plays the latter role. 5) DAG and MAG may activate TRP and TRPL by a mechanism that has not been established. A signaling complex that includes TRP and PLC is coordinated by INAD. 6) Na+/Ca2+ exchanger channel (Calx) pumps Ca2+ out of the photoreceptor cell. 7) Arrestin 1 and 2 bind rhodopsin to terminate the cascade with Arrestin 2 as the dominant arrestin in both D. melanogaster and butterflies. INAD and NinaC bind to each other, and both bind calmodulin, to accelerate arrestin binding rhodopsin. STOPS is another protein that terminates phototransduction. Black stars signify differences in phototransduction between D. melanogaster and Lepidoptera due to gene duplication. White stars represent differences in relative expression of UnRh, Rh7, and Calx between moths and butterflies, and in trp and trpl between flies, moths, and butterflies. Further description of these genes can be found in supplementary Table S11, Supplementary Material online.
