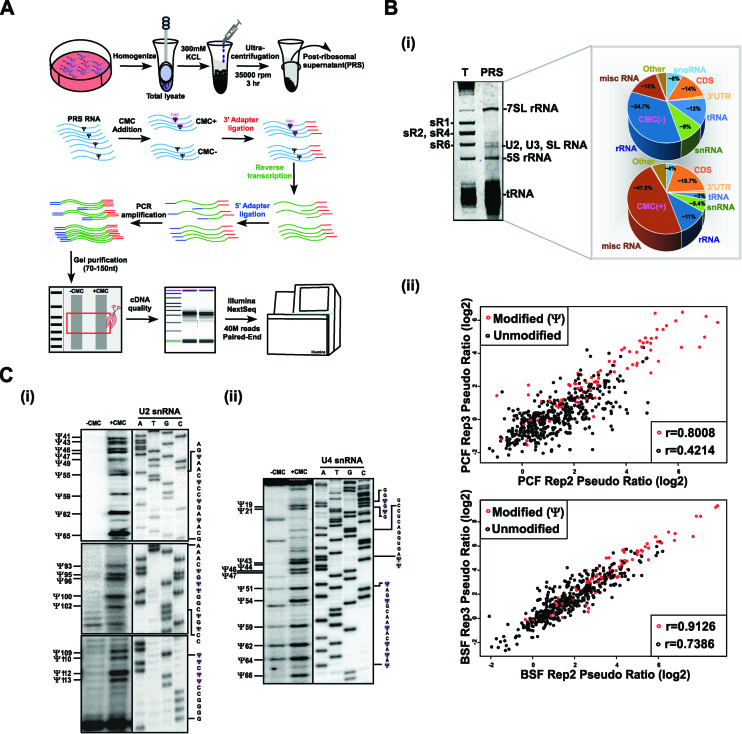
Figure 1.
Small RNA Ψ-seq in T. brucei. (A) Schematic presentation of the small RNA Ψ-seq protocol. The scheme illustrates the extract preparation, fractionation, and the small RNA Ψ-seq methodology. (B) Enrichment of Ψs in small RNA Ψ-seq. Whole-cell extracts from 2 × 109 PCF and BSF cells was prepared and depleted of ribosomes; RNA (800ng) was subjected to CMC treatment and used to prepare small RNA libraries, as described in (A). (i) Enrichment of small RNAs in PRS. RNA was separated on a 10% denaturing gel and stained with ethidium bromide. (T) total RNA; PRS. The identity of each RNA type is indicated. The pie diagram represents the types of RNA present in the Ψ-seq library. (ii) The reproducibility of pseudouridines in Ψ-seq libraries. Pairwise comparison of Ψ-fc(log2) from two libraries showing pseudouridylated sites (in red) and non-modified sites (in black) in snRNA. Pearson's correlation coefficient (r) is indicated. Fourteen independent biological replicates were used to detect Ψs in small RNA Ψ-seq. (C) Validation of Ψs in T. brucei U2 and U4 snRNA. Total RNA (100μg) treated with CMC (+CMC) or untreated (–CMC) was subjected to primer extension with region specific primers and analyzed on a 12% polyacrylamide gel (7M urea). The results along with DNA sequencing performed using the same primer are presented for U2 (i) and U4 (ii) snRNA. The position of the Ψs are indicated (one nt after the actual stop seen in the gel), as well as the RNA sequence. Contrast adjusted blots are separated by bold lines.