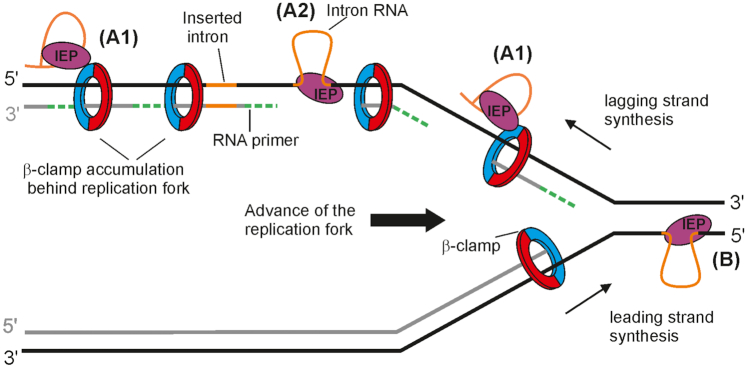
Figure 7.
Possible model for mobility of group II introns displaying a replication orientation bias. A1 and A2 represent the pathway used by RmInt1 (En- intron) and probably for the retrotransposition of En+ introns, which show a bias for insertion into the lagging strand template. They localize to ssDNA by interacting with g that the intera-clamp; it could take place with stalled replication forks, during the shifting between different host-encoded DNA polymerases or with the DnaN that remains bound to DNA in the lagging strand behind the replication fork (A1) and then reverse splice into its ssDNA target site (A2); DNA polymerase would disengage upon encountering the highly structured intron or tightly bound IEP, leaving the nascent DNA strand in position to be used as a primer by the IEP to reverse transcribe the inserted intron RNA. A host DNA polymerase probably resumes DNA synthesis after reverse transcription of the intron RNA. (B) It represents the pathway used by mutants of En+ introns lacking endonuclease activity and displaying an orientation replication bias toward use of the nascent leading strand DNA as a primer for reverse transcription of the inserted intron RNA. After reverse splicing into the leading strand template without cleavage of the opposite strand, the intron RNP is in a position to use the nascent leading strand from an approaching replication fork as a primer.