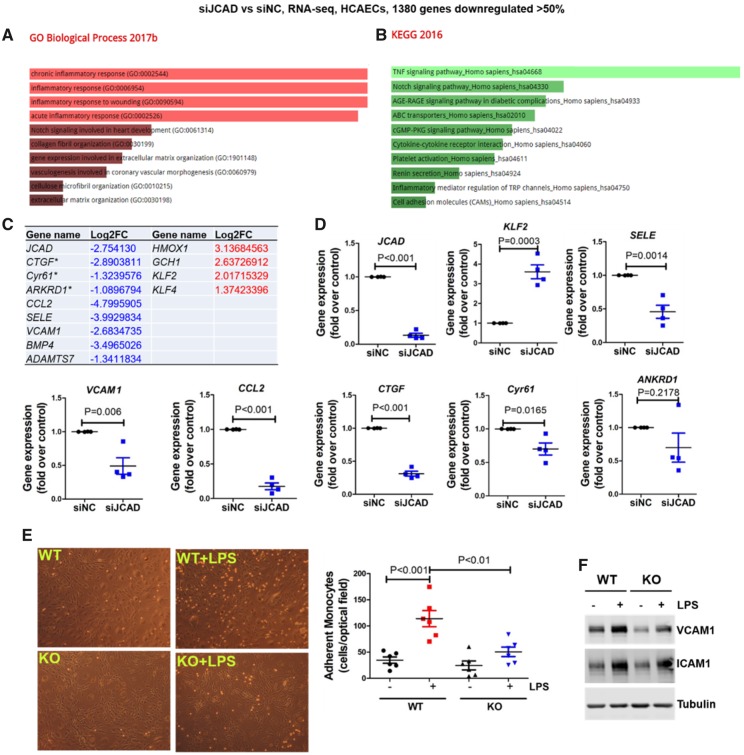
Figure 5.
JCAD deficiency attenuates vascular inflammation by acquiring an atheroprotective transcriptional program. (A–B) Gene ontology (A) and KEGG2016 pathway (B) analysis of differentially expressed genes in JCAD depleted HCAECs by RNA-seq. RNA-seq data were deposited in NCBI Gene Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number of GEO: GSE102498. (C) Selected atherosclerosis-relevant genes regulated by JCAD revealed by RNA-seq. * indicates established downstream target genes of YAP/TAZ. For a whole list of differentially expressed genes, please refer to Supplementary material online, Table S7. (D) Validation of RNA-seq data by real time PCR. Select downregulated and upregulated genes were presented, n = 4. (E–F) JCAD deficiency attenuates LPS-induced vascular inflammation. JCAD+/+ (WT) and JCAD−/− (KO) primary mouse lung endothelial cells were stimulated with LPS for 6 h before monocyte (THP1) adhesion assay was performed (E, n = 6). In parallel, whole cell lysates were collected for Western blot analysis to detect protein expression of VCAM1 and ICAM1 (F, n = 4), ***P < 0.001 vs. WT, ##P < 0.01 vs WT + LPS. Log2FC, Log2 fold change; siNC, scramble control siRNA; siJCAD, JCAD siRNA; LPS, lipopolysaccharide.