Figure 4.
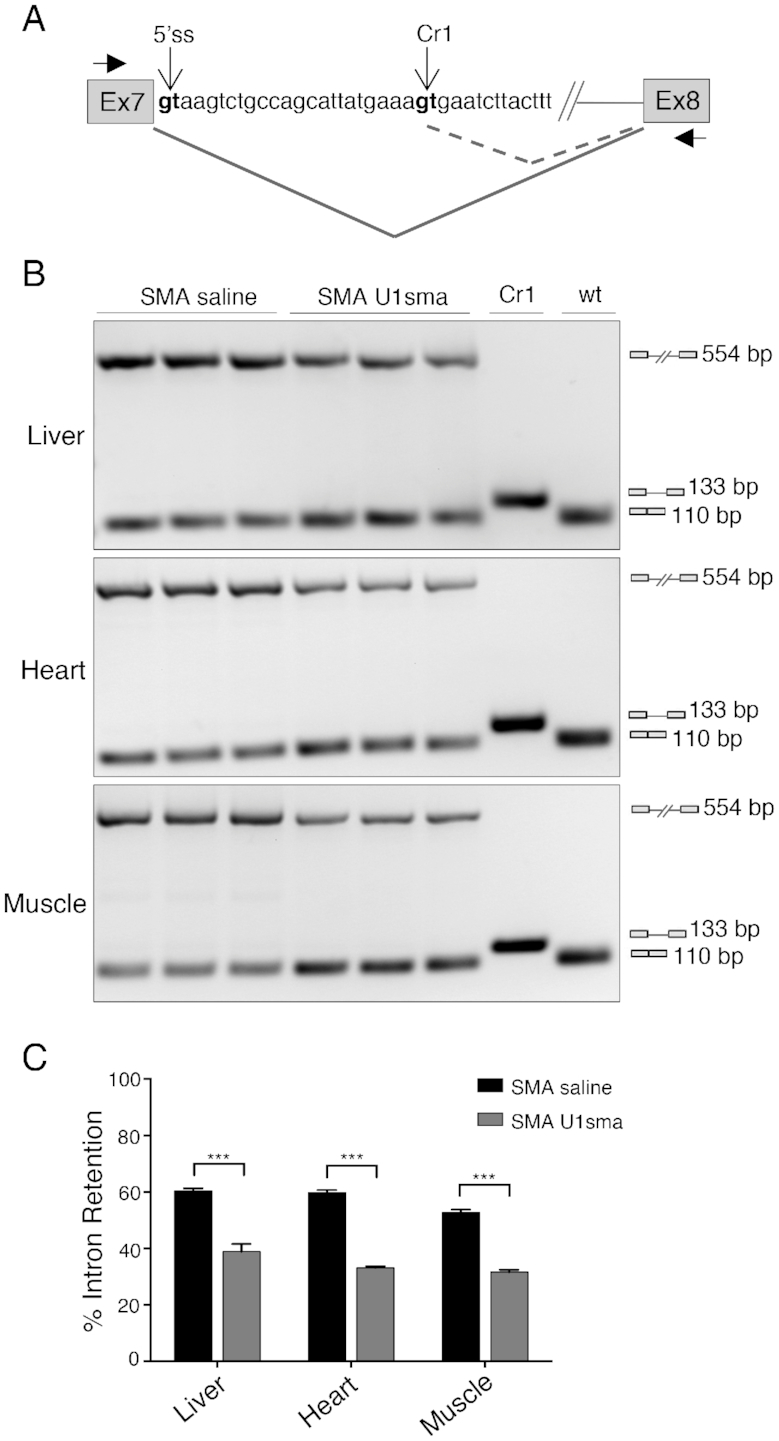
AAV9-ExSpeU1sma in vivo does not induce mis-splicing of SMN2 intron 7. (A) Schematic representation of SMN2 exon 7-intron 7-exon 8 structure. The wild-type (5′ss) and cryptic donor sites identified in (50) (Cr1), as well as the position of the primers (Ex7 and Ex8) used in the amplification experiments are indicated. Differential splicing events are shown. (B) SMA severe mice (smn−/−, SMN22TG/0) treated with saline (n = 3; P7) and AAV9-ExSpeU1sma (n = 3; P7) were analyzed by endpoint PCR with Ex7 and Ex8 primers. DNase-treated RNA samples correspond to samples analyzed in Figure 3C (P7 bars) and Supplementary Figure S4A (P7 lanes). Amplification experiments in SMA saline and SMA U1sma samples showed an upper (554 bp) and a lower (110 bp) band that corresponds to retained intron 7 and correctly spliced transcripts, respectively. Cr1 and wt lanes are control amplifications of plasmids that contain the spliced cryptic (133bp) or the normal 5′ss (110 bp) sequences, respectively. Amplification experiments of AAV9-ExSpeU1sma-treated samples did not show any Cr1 133bp-long band. (C) Histogram shows quantification of the percentage of intron 7 retention. Data are expressed as mean ± SD of two technical replicates on three mice and statistical analysis was performed using Student’s t-test (***P < 0.001).
