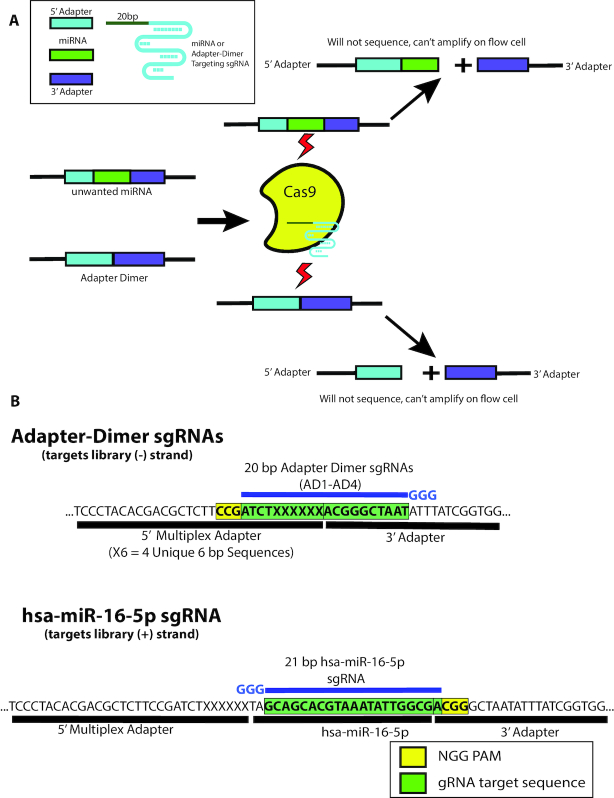
Figure 1.
Depletion of adapter dimer and overabundant miRNAs with MAD-DASH. (A) MAD-DASH employs Cas9 and adapter dimer- or miRNA-specific sgRNAs to selectively deplete these sequences from final libraries. Cleaved sequences will not amplify during the second round of PCR amplification, and are also no longer suitable substrates for bridge amplification during Illumina sequencing (though they will still be able to bind to the flow-cell using the P5/P7 sequences remaining on each cleaved library). (B) Design of adapter dimer and hsa-miR-16–5p sgRNAs using available pre-existing PAM sites on our smRNA-seq 5′- and 3′-adapters. Adapter dimer targeting sgRNAs use a minus strand ‘NGG’ and have 10 bp of homology to both the 5′- and 3′ adapter, ensuring target cleavage specificity for adapter dimer sequences. hsa-miR-16–5p uses a 3′ PAM site one base away from the 3′—adapter ligation junction, which can be generalized to other smRNAs and provides highly specific targeting while minimizing off-targets. Shown are only the plus strand of non-PCR tailed sequences. Green boxes indicate the plus strand sequence corresponding to sgRNA location in the dsDNA library, while the yellow box indicates the plus strand location of the ‘NGG’ PAM in the dsDNA library. sgRNA location is depicted in blue along with the three 5′ ‘G’ nucleotides necessary for high levels of T7 in vitro transcription.