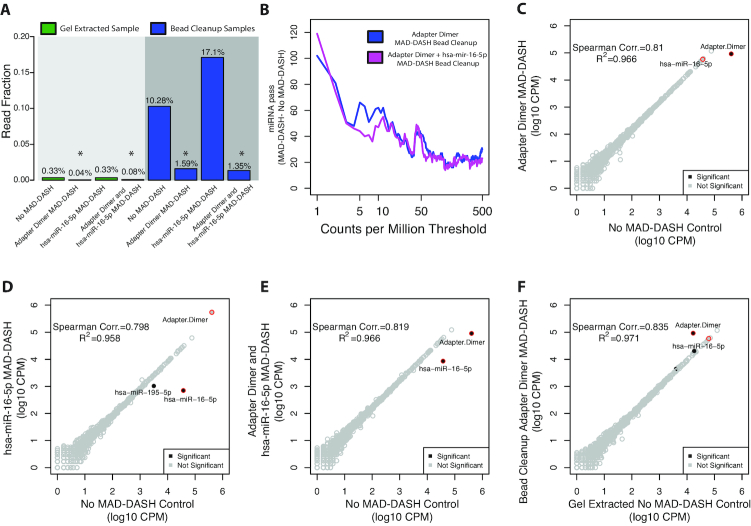
Figure 2.
MAD-DASH smRNA-seq reduces adapter dimer and hsa-miR-16–5p in human brain total RNA samples. (A) Averaged read fraction of downsampled and CPM normalized adapter dimer sequences in gel extracted and bead cleanup samples. MAD-DASH in bead cleanup samples significantly depletes adapter dimer individually and in combination with depletion of hsa-miR-16–5p simultaneously. Asterisks indicate that a given read fraction was determined to be significantly altered compare to the respective cleanup's no MAD-DASH control using DESeq2 (FDR < 0.05). (B) Plot depicting the difference in the number of mature miRNA species reaching a specified count per million threshold between bead cleanup adapter dimer MAD-DASH smRNA-seq replicates and MAD-DASH control replicates. Downsampling and CPM normalization was performed as described in ‘Materials and Methods’ section. (C–F) Read counts from normalized replicate groups for bead cleanup treated versus control MAD-DASH samples targeting (C) adapter dimer (D) hsa-miR-16–5p and (E) adapter dimer and hsa-miR-16–5p (F) adapter dimer compared to gel extracted no-Cas9/sgRNA control. Significantly different sequences are filled black circles, with those having a log2-fold-change > 1 being labeled with text. Non-significantly different miRNAs are depicted as open gray circles. Adapter dimer and hsa-miR-16–5p are depicted as red circles with the same gray or black fill to denote significance. Significance was determined with DESeq2 and set as a Benjamini–Hochberg corrected P-value < 0.05.