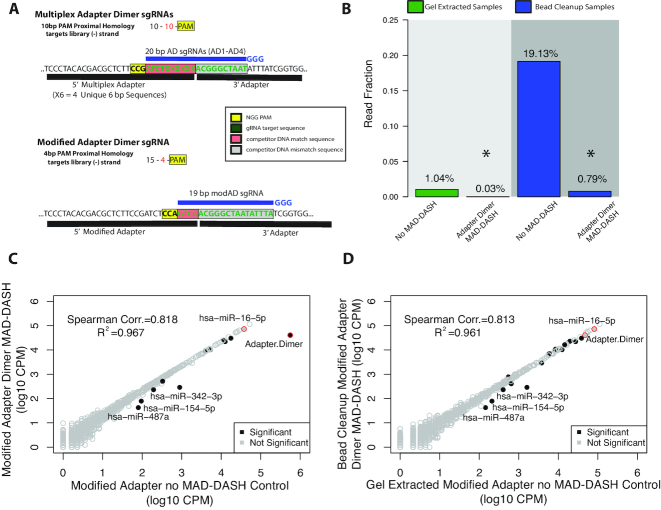
Figure 3.
Rational design of a modified 5′ adapter with alternate PAM site enhances depletion of adapter dimer with MAD-DASH. (A) Design of the modified 5′ adapter compared to the multiplex 5′ adapter. Compared to the 10 bp competitor mismatch—10 bp competitor match—PAM design used in our first MAD-DASH iteration, the modified adapter uses 15 bp competitor mismatch—4 bp competitor match—PAM design and is predicted to have as much as 100-fold less binding to other non-target sequences in the library. Shown are the plus strands of non-PCR tailed sequences. Red boxes indicate the plus strand sequence corresponding to competitor match sequence in the dsDNA library, while the gray box indicates the sequence corresponding to sequence that drives adapter dimer target specificity, i.e. competitor mismatch sequence. The yellow box indicates the plus strand location of the ‘NGG’ PAM in the dsDNA library. sgRNA location is depicted in blue along with the three 5′ ‘G’ nucleotides necessary for high levels of T7 in vitro transcription. (B) MAD-DASH smRNA-seq using the modified 5′ adapter demonstrates substantially greater depletion of adapter dimer in bead cleanup samples than when using the multiplex adapter, and achieves a lower average read fraction than in modified adapter MAD-DASH control gel extraction samples. Normalized read fraction of downsampled and CPM normalized adapter dimer sequences for both adapter strategies are depicted. Reductions of adapter dimer in both gel extracted and bead cleanup samples were significant (DESeq2 adjusted P-value < 0.05). (C and D) Read counts from normalized replicate treated vs control groups prepared using the modified 5′ adapter. Bead cleanup MAD-DASH samples depleted of (C) modified adapter dimer and (D) bead cleanup modified adapter dimer compared to gel extracted no-Cas9/sgRNA modified adapter control are shown. Significantly different sequences are filled black circles, with those having a log2-fold-change > 1 being labeled with text. Non-significantly different miRNAs are depicted as open gray circles. Adapter dimer and hsa-miR-16–5p are depicted as red circles with the same grey or black fill to denote significance. Significance was determined with DESeq2 and set as a Benjamini–Hochberg corrected P-value < 0.05