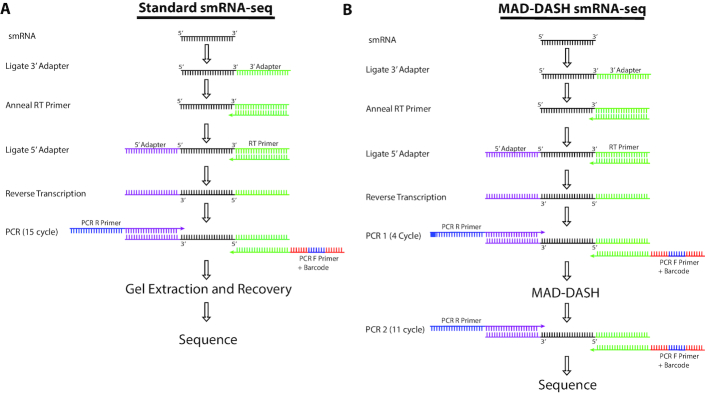
Figure 5.
Illustration of MAD-DASH and standard smRNA-seq workflows. While MAD-DASH involves an extra PCR amplification in addition to the MAD-DASH CRISPR/Cas9 in vitro digestion, the extra ∼3 h to accomplish this relative to finishing the PCR in our standard protocol are more than offset by no longer needing to perform gel extraction, DNA recovery and library concentration. For nearly a hundred samples, we estimate that researcher time for smRNA-seq library construction can be reduced to only 1 day using MAD-DASH smRNA-seq and that this throughput can be further improved with greater protocol familiarity or automation.