Figure 5.
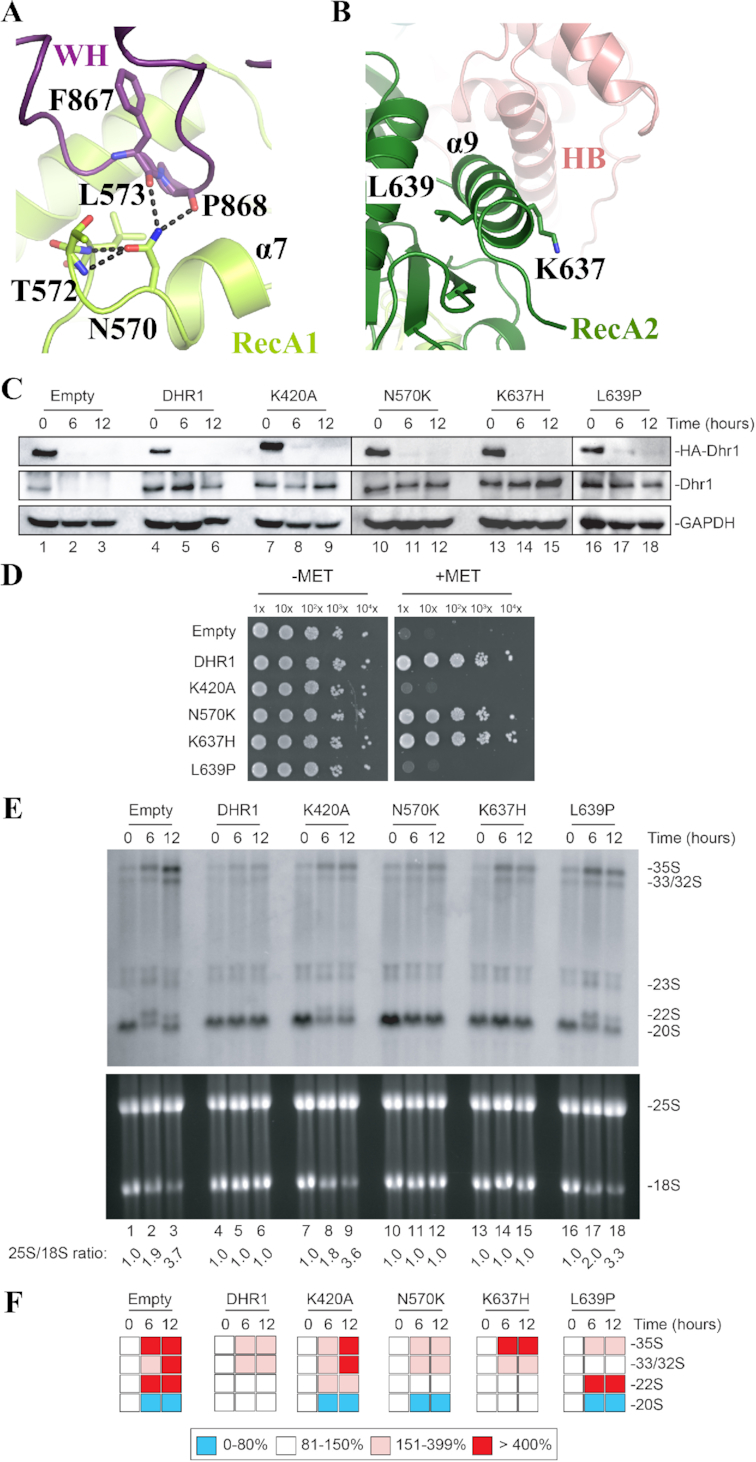
Functional analysis of DHX37 mutations associated with diseases. (A) The RecA1 mutation N570K (equivalent to N419 in human DHX37) is expected to disrupt the compact hydrogen bonding network (dashed black lines) normally formed between this residue and its neighbors. See also Figure 2B to localize the mutation in the global structure of the protein. (B) The RecA2 mutations K637H (R487 in human) and L639P (L489 in zebrafish) are located close to each other in helix α9. K367 is solvent exposed and positively charged, L639 is in the center of the helix and its substitution by a Proline is expected to disrupt the helix. (C) All four Dhr1 mutations are stably expressed. Western blot analysis showing the progressive extinction of expression of endogenous HA-Dhr1, specifically detected with an anti-HA antibody (top panels), and the expression of the complementing wild-type Dhr1 and four mutants (middle panels, detection with an anti-Dhr1 antibody). As loading control, blots were probed for GAPDH. The experiments in panels C and E are showing a time course of transfer from a medium lacking methionine (0-h time points) to a medium containing methionine (6- and 12-h time points). (D) Mutations K420A and L639P are deleterious for growth. Serial dilutions of the indicated yeast strains were incubated on agar plates for 3 days at 30°C in the absence (–MET) or presence (+MET) of methionine. (E) Mutations K420A and L639P are deleterious for ribosome biogenesis. Top, Northern blot analysis detecting major pre-rRNA species (indicated to the right). The probe used was specific to internal transcribed spacer 1 (ITS1) sequence (see materials and methods). Bottom, ethidium bromide-stained denaturing agarose gel showing the amounts of 18S and 25S rRNAs. The 25S/18S ratio established from bioanalyzer electropherogramms is indicated below the gels. (F) Quantification of panel E. Pre-rRNA processing intermediates were quantified with a Phosphor imager. Results are shown as a heat-map.
