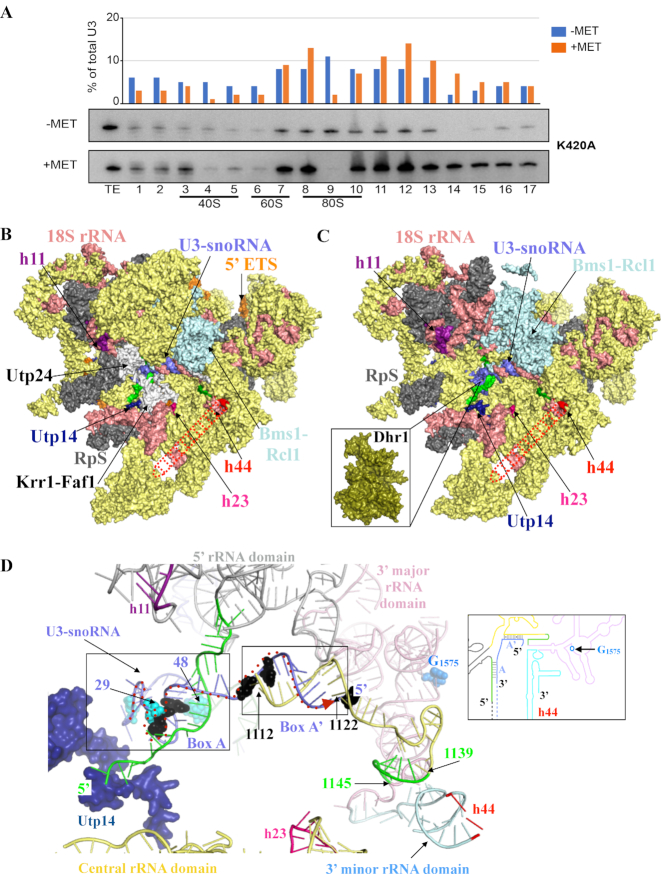
Figure 7.
The involvement of Dhr1 in CPK formation and pre-90S assembly. (A) Velocity gradient analysis showing that in the absence of Dhr1′ catalytic activity, the box C/D snoRNA U3 remains bound to pre-90S ribosomes. Total cell extract from yeast cells expressing the K420A mutation in the presence of wild-type Dhr1 (–MET) or in its absence (+MET, see legend to Figure 4 for details) were separated on 5–50% sucrose gradients. RNA extracted from each collected fraction was resolved on denaturing polyacrylamide gels and analyzed by northern blotting with a probe specific to U3. The U3 signal detected in each lane was quantitated with a Phosphor imager and expressed as a percentage of total U3 signal (histogram in the upper part of the panel). For reference, positions of 40S, 60S, and 80S ribosomal peaks (as established by OD260) are indicated. TE, total extract control sample. It is not clear at this stage why U3 appears more abundant in the +MET panel (since the same amounts of total cell extracts were loaded on each -MET and +MET panels, but we note that an identical observation was made independently with the same K420A mutation (see Figure 2E in (16)). This experiment was repeated three times, a representative example is shown. (B) Surface representation of S. cerevisiae pre-90S particle as determined by cryo-EM at 3.8 Å overall resolution (58). Assembly factors modeled are colored in yellow and ribosomal proteins (RpS) in gray. For the sake of simplicity, the factors and RpS are not named on the figure, but their identify is known (see PDB 5WLC). The three rRNA helices with which Dhr1 interacts are shown: h11 (purple), h23 (pink) and h44 (red dashed line, this latter helix is largely undefined in the cryo-EM map likely due to high intrinsic flexibility). Nucleotides from pre-18S rRNA forming the CPK in mature 18S are colored in green in panels B to D (see also Figure 1); U3 is in blue. Selected assembly factors are highlighted (Utp14, Utp24-Krr1-Faf1, Bms1-Rcl1). The 18S rRNA is in salmon and the 5′ ETS in orange. (C) Simplified version of panel B (excluding factors not found in association with Dhr1-K420A in (16)). At this stage, Dhr1 has not been mapped on cryoEM structures because it was absent from the purified precursor ribosomes analyzed. Inset: surface representation of the Dhr1 helicase module at the same scale as pre-90S pointing toward its site of action: the U3 snoRNA (in blue) on the precursor small subunit. This is where we suggest Dhr1 binds after the release of the assembly factors Utp24-Krr1-Faf1 (see text for details). (D) ‘At the heart of the SSU-processome’: representation of RNA duplexes formed between the 5′ end of U3 and the 18S rRNA. The Dhr1-interacting protein Utp14 is colored in dark blue. U3 nucleotides identified as interacting directly with Dhr1 in in vivo cross linking studies, or as suppressing the growth defect of a cryo-sensitive dhr1 allele, are shown in cyan and black spheres, respectively (16). CPK formation is essential to small ribosomal subunit biogenesis because it brings together sequences from the four rRNA domains which are far apart in the primary structure. The four rRNA domains are colored as follows: 5′ domain (light grey), central domain (yellow), 3′ major domain (pink), and 3′ minor domain (light blue) according to the color scheme used in Figure 1 and in the inset. The duplexes formed between the U3 boxes A and A’ and the 18S rRNA are highlighted in black boxes. The G1575 nucleotide, which is N7-methylated by the Bud23-Trm112 complex is shown as blue spheres. The proposed path of the Dhr1 helicase onto the U3 snoRNA-pre-rRNA duplexes during unwinding is depicted as a dashed red arrow from 3′ to 5′ end (left to right). Inset: Cartoon representation of the 18S rRNA precursor bound to U3, using the same color code as in the 3D model representation.