Figure 2.
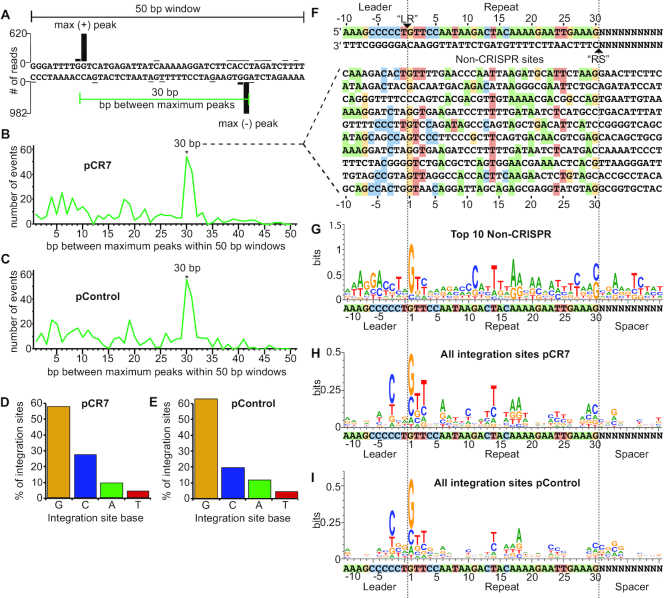
Non-CRISPR integration sites resemble CRISPR repeats. (A) Spacing between maximum integration peaks on plasmid tracks shown in Figure 1 were measured within 500 random 50 bp windows. (B, C) The number of times each bp spacing between maximum peaks was counted on pCR7 (B) and pControl (C). (D, E) Percent of total integration sites located at guanine, cytosine, adenine, and thymine on pCR7 (D) and pControl (E). (F) The top ten 30 bp non-CRISPR sites on pCR7. List includes any sequences that had more than 40 reads at either the ‘LR’ or ‘RS’ location of the sequence. Homology is highlighted for each nucleotide that matches the CRISPR repeat. (G) WebLogo of non-CRISPR sequences listed in F. (H, I) WebLogo of all integration events on pCR7 (H) and pControl (I).