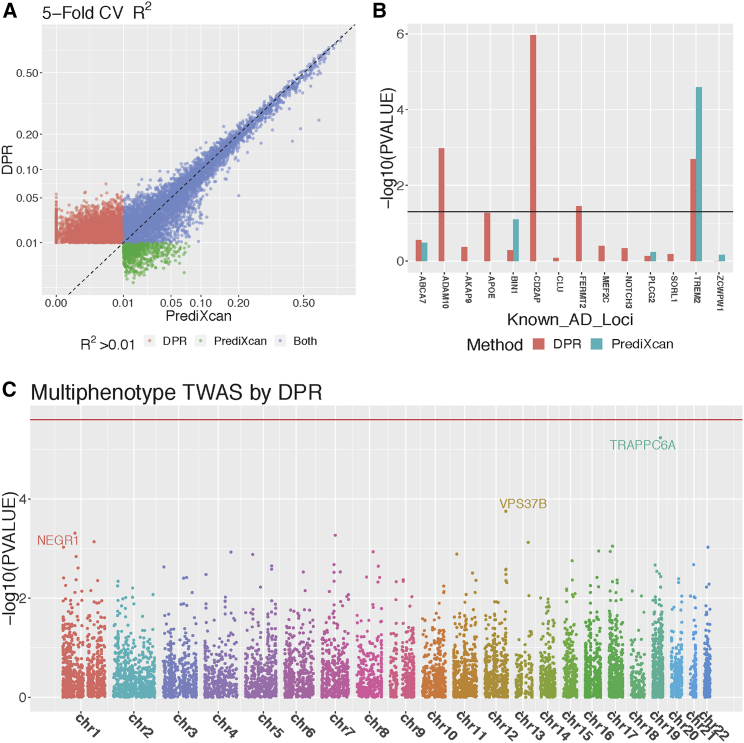
Figure 3.
TWAS Results of Studying Alzheimer's Disease
Transcriptome-wide 5-fold cross validation R2 (A) by PrediXcan and DPR with 499 ROS/MAP training samples, with different colors denoting whether the imputation R2 > 0.01 by DPR, PrediXcan, or both methods (genes with R2 > 0.01 by both DPR and PrediXcan were excluded from the plot). TWAS results (B) at known AD loci using GWAS summary-level statistics from IGAP and imputation models fitted from ROS/MAP data, where missing values are due to NULL imputation models by PrediXcan. Manhattan plot (C) for the multiphenotype TWAS (with neurofibrillary tangle density and -amyloid load), using individual-level ROS/MAP data.