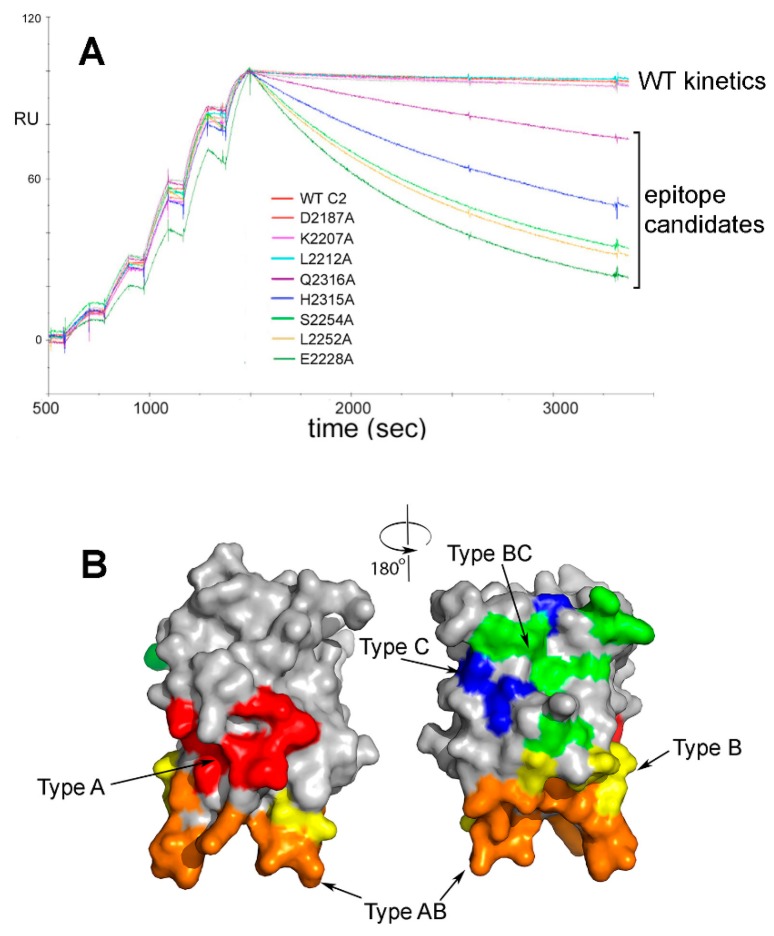
Figure 1.
Surface plasmon resonance (SPR)-based epitope mapping. (A). Representative superimposed sensorgrams showing single-cycle kinetics experiments in which rFVIII-C2 protein and rFVIII-C2 variants with a single surface-exposed side chain mutated to alanine were injected at increasing concentrations over a FVIII-specific MAb captured on a biosensor [97]. Residues were flagged as potential contributors to the epitope if the kd for the FVIII-C2 mutein was >2.0X the kd for the wild-type rFVIII-C2 protein. Alanine substitutions at residues E2228, L2252, S2254, H2315 and Q2316 met this criterion in this set of experiments with MAb 1B5. Separate SPR runs (not shown here) identified residues F2196, T2197, N2198, F2200, T2202, R2220, Q2222, N2225 and K2239 as also possibly contributing to the epitope recognized by this MAb. (B). Front and back views (rotated 180°) of the FVIII C2 domain crystal structure [100], with surfaces colored to indicate the 5 partially-overlapping B-cell epitopes recognized by 11 neutralizing MAbs. The MAbs and their cognate epitopes were designated Types A, AB, B, BC and C on the basis of competition ELISA experiments. These MAbs inhibit distinct binding interactions and functions of FVIII, e.g., binding to negatively charged phospholipid surfaces, von Willebrand factor, and other proteins comprising the ‘intrinsic tenase’ complex [88]. Type A: red; Type AB: orange; Type B: yellow; Type BC: green; Type C: blue.