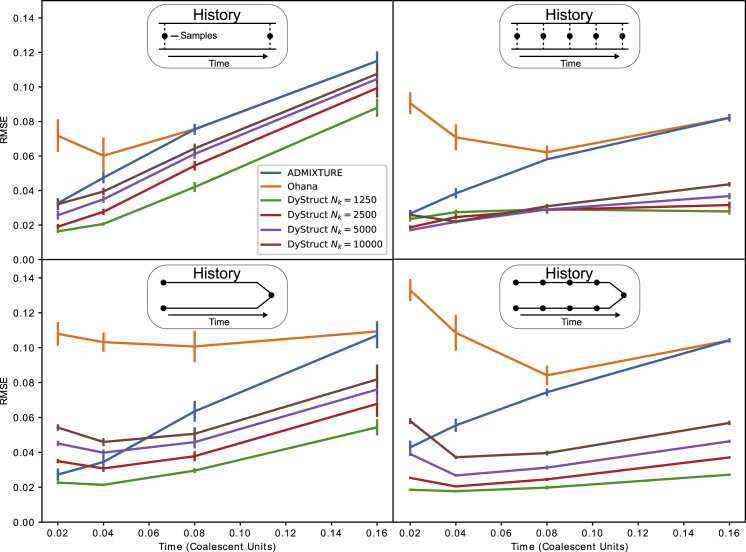
Figure 2.
Performance of DyStruct, ADMIXTURE, and Ohana on Simulated Data for Two Historical Scenarios and Two Sampling Schemes
Top: individuals are mixtures of two ancestral populations (the “mixture” scenario). Bottom: ancient individuals are sampled solely from one of two ancestral populations that merge at present, and modern samples from the present-day admixed population (the “merger” scenario). Models were compared by computing the root-mean-square-error (RMSE) on ancestry estimates between the simulated ground truth and the output from both models. Diagrams above each plot show the historical scenario simulated, and black circles denote approximately when individuals are sampled. The x axis displays total simulation time between the first and last sample (1 coalescent unit = 2N generations for simulated N = 2,500). The y axis gives the mean RMSE across models (vertical lines denote 1 standard error estimated from 10 replicates). DyStruct was run with varying population sizes to explore model performance when the true size is unknown. DyStruct’s ancestry estimates improve when individuals are more densely sampled in time, while ADMIXTURE’s remain similar. Both models outperform Ohana.