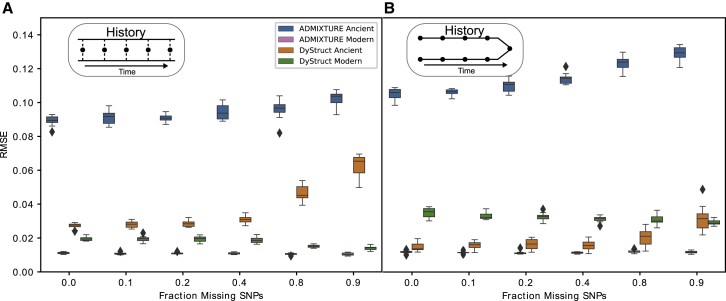
Figure 4.
DyStruct and ADMIXTURE’s Performance on Sparse Pseudo-haploid Samples
Boxplots of root mean square error (y axis) between the ground truth ancestry and model estimates for ancient and modern samples. The fraction of missing SNPs varied across ancient samples (x axis), while modern samples had full data. Ancient samples are psuedo-haploid, where only a single allele is observed at each locus and encoded by 0 or 2. DyStruct’s ancestry estimates on ancient samples are more accurate than ADMIXTURE across degrees of missing data.