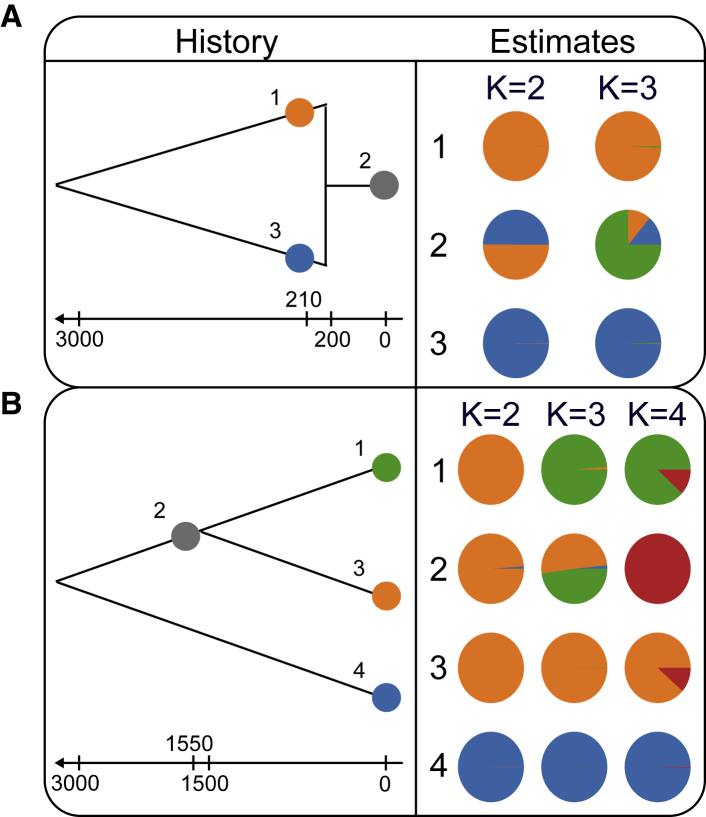
Figure 5.
DyStruct’s Performance on Coalescent Simulations
Diagrams on the left depict the historical scenario simulated. Colored circles denote where in the population phylogeny individuals were sampled. Arrows and notches below each diagram give the generation time of sampled individuals and historical events. Pie charts on the right give the mean ancestry estimates for each population across 10 replicates.
(A) At K = 2, DyStruct can accurately detect the contribution of the ancient populations to the modern one. Ancient samples are assigned nearly a single ancestry component (color), while modern samples appear as admixed between ancient populations. At K = 3, DyStruct assigns a new cluster to the modern population, but still identifies shared ancestry from its ancestral contributors.
(B) At K = 2, DyStruct identifies the oldest split in the phylogeny. At K = 3, DyStruct splits the ancient population by clusters corresponding to its two descendants. At K = 4, the ancient population gets its own cluster, but DyStruct still identifies shared ancestry between it and its two descendant populations.