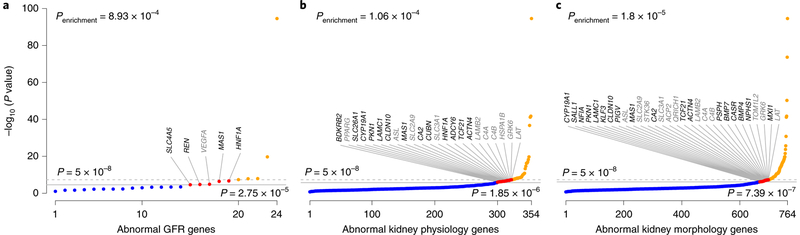
Fig. 3 |. Human orthologs of genes with renal phenotypes in genetically manipulated mice are enriched for association signals with eGFR.
a-c, Signals in candidate genes identified on the basis of the mouse phenotypes of abnormal GFR (a), abnormal kidney physiology (b) and abnormal kidney morphology (c). The y axis shows −log10 (P) for association with eGFR in the trans-ancestry meta-analysis for the variant with the lowest P value in each candidate gene. The dashed line corresponds to genome-wide significance (P = 5×10−8), and the solid gray line corresponds to the experiment-wide significance threshold for each nested candidate gene analysis. Orange, genome-wide significance; red, experiment-wide but not genome-wide significance; blue, no significantly associated SNPs. Genes are labeled if they reached experiment- but not genome-wide significance; black font indicates genes not mapping to loci reported in the main analysis. Enrichment P values correspond to the observed number of genes with association signals below the experiment-wide threshold against the number expected on the basis of the complementary cumulative binomial distribution (Methods).