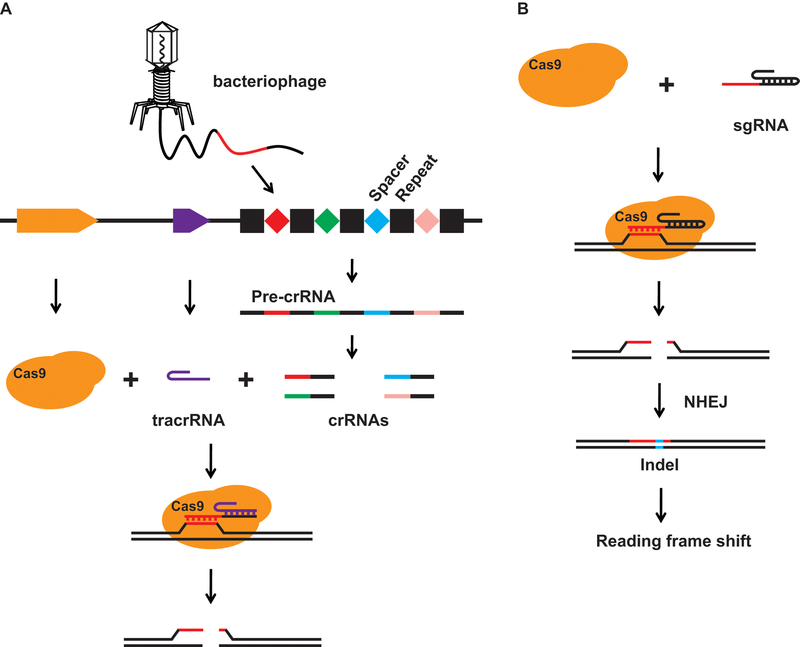
Figure 1. Natural and engineered CRISPR systems.
(A) Schematic of a natural CRISPR pathway. Foreign DNA is captured and inserted between repeats in a CRISPR locus in a bacteria genome. CRISPR array is transcribed and then processed into multiple crRNAs, each carrying a single spacer sequence and a repeat sequence. The crRNA forms a complex with Cas9 and crRNA and directs the complex to the foreign DNA carrying the same sequence as the spacer. Cas9 then generates a double strand break on the foreign DNA. (B) Schematic of an engineered Cas9 genome editing tool. It comprises a Cas9 endonuclease and a single guide RNA (sgRNA). Cas9 and sgRNA form a complex. The guide sequence on sgRNA directs the complex to the target site. Cas9 generates a double strand break, which is repaired via the error-prone non-homologous end-joining (NHEJ), leaving a small insertion or deletion (Indel) at the target site. Indels in protein coding region cause reading frame shift and early termination of protein translation, resulting in loss of protein expression.