Abstract
The RNA polymerase II complex (pol II) is responsible for transcription of all ∼21,000 human protein-encoding genes. Here, we describe sixteen individuals harboring de novo heterozygous variants in POLR2A, encoding RPB1, the largest subunit of pol II. An iterative approach combining structural evaluation and mass spectrometry analyses, the use of S. cerevisiae as a model system, and the assessment of cell viability in HeLa cells allowed us to classify eleven variants as probably disease-causing and four variants as possibly disease-causing. The significance of one variant remains unresolved. By quantification of phenotypic severity, we could distinguish mild and severe phenotypic consequences of the disease-causing variants. Missense variants expected to exert only mild structural effects led to a malfunctioning pol II enzyme, thereby inducing a dominant-negative effect on gene transcription. Intriguingly, individuals carrying these variants presented with a severe phenotype dominated by profound infantile-onset hypotonia and developmental delay. Conversely, individuals carrying variants expected to result in complete loss of function, thus reduced levels of functional pol II from the normal allele, exhibited the mildest phenotypes. We conclude that subtle variants that are central in functionally important domains of POLR2A cause a neurodevelopmental syndrome characterized by profound infantile-onset hypotonia and developmental delay through a dominant-negative effect on pol-II-mediated transcription of DNA.
Keywords: POLR2A, RPB1, RNA polymerase II complex, de novo variants, infantile-onset hypotonia, neurodevelopmental syndrome, dominant-negative effect, haplo-insufficiency, desert regions, desert Z score
Introduction
Human POLR2A (MIM: 180660) encodes the highly-conserved RPB1 protein, which is the largest of twelve subunits of the essential RNA polymerase II (pol II) enzyme.1, 2 This protein complex is responsible for the transcription of all protein-encoding genes, as well as several long and short non-coding RNA genes.3 Due to its central role in gene expression, pol II and the regulation of its activity have been studied in depth since its discovery fifty years ago.4 Biochemical and structural studies have shown that transcription initiation by pol II requires a set of basal (or general) transcription factors. After assembly of the pol II initiation complex at the promoter region, these factors are exchanged for transcription elongation factors including TFIIS, DSIF, and P-TEFb to allow processive RNA synthesis by pol II.5 Detailed structural analysis of the pol II of Saccharomyces cerevisiae (S. cerevisiae)6 revealed different aspects of RNA synthesis, including initiation,7, 8, 9, 10 nucleotide binding,11 chain elongation,12, 13, 14, 15, 16 error correction, and back tracking.17, 18, 19, 20 For example, during the elongation cycle, the incorporation of the incoming nucleotide involves movement of the so-called RPB1 trigger loop, which induces the forward movement of pol II over the DNA template strand coincident with nucleotide selection. In vivo dynamics of pol II during initiation and promotor pausing were studied in GFP-RPB1 knock-in cells, and this research demonstrated that the continuous release and reinitiation of promotor-bound pol II is important for transcriptional regulation.21 Mutational studies focusing on S. cerevisiae and human RPB1 indicated that residues in the trigger loop region are controlling the rate of RNA synthesis.22, 23 Experiments in S. cerevisiae indicated that the phenotypic consequences of rpb1 variants in yeast might be concealed by transcript buffering24 and might only become apparent under conditions of environmental or nutrient stress.25 Experiments in Arabidopsis mutants that encode truncated RPB1 with different sizes of shortened C-terminal domain (CTD) show disturbed cell cycling control, demonstrating its importance in transcriptional regulation of cell cycle genes.26 Despite its critical role in transcription, POLR2A has not been implicated in human disease thus far.
Here, we describe 16 individuals harboring de novo heterozygous genetic variants in POLR2A. We acknowledge that, although the occurrence of de novo variants provides an important clue toward variant pathogenicity,27 this is not decisive28 because large-scale sequencing of individuals without severe pediatric disease has revealed that protein-changing genetic variants are present in all ∼21,000 protein-encoding genes.29 Similarly, as individuals carrying a variant in a certain gene are increasingly found through gene-matching initiatives,30 phenotypic overlap might merely reflect the general characteristics of individuals undergoing whole-exome or whole-genome sequencing (WES, WGS). In addition, neither the presence nor the absence of evidence from functional analyses can be used to prove or refute pathogenicity. This transition of genetics from a dichotomous field to an exciting but puzzling world full of shades of gray also offers new avenues of study. Deep phenotyping analyses might help to accurately expose the extent of the phenotypic overlap to support pathogenicity and so might severity metrics from population genetics. We here apply an iterative process wherein we combine several lines of evidence, including a detailed phenotypic analysis, an assessment of variant severity metrics, and structural biology and functional analyses in both S. cerevisiae and HeLa cells to assess the pathogenicity of the 16 identified variants in POLR2A in detail.
Material and Methods
Phenotypes of Affected Individuals
The cohort was assembled through the GeneMatcher initiative.30 The inclusion criterion was the detection of a de novo heterozygous variant in POLR2A (GenBank: NM_000937.4) confirmed by either clinical or research trio WES or WGS or by confirming the exclusion of the heterozygous variant in both parents. Physicians first provided a detailed phenotypic description of the individual. In a second round, questions were asked regarding the specific clinical features of the individual to further define the clinical overlap among individuals. Additionally, doctors and parents together performed a thorough evaluation of the individuals’ attained milestones by using items of the Denver Scale.31 We used these data to assess both the rate of development over time and developmental-domain-specific delays. To assess these focus points, we calculated Z scores. The Denver Scale provides information on the fiftieth (p50) and ninetieth (p90) percentiles of the age in months, at which the normal population attains developmental milestones. The ninetieth percentile corresponds to 1.28 standard deviations (SDs). SD was calculated by (p90 – p50)/1.28. Z scores were calculated as (x – p50)/SD, wherein x is the month the individual attained the developmental milestone. We performed the statistical analysis in R programming language by using log10 transformation. All procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2000. The legal guardians for all individuals agreed to the participation of their child in the study and signed the appropriate consent forms, in agreement with institutional and national legislation for the different centers. Permission for publication of pictures was given separately.
Metrics of Variant Severity
Probability of intolerance to loss of function (pLI) scores for loss-of-function variants and Z scores for synonymous and missense variants were retrieved from the Genome Aggregation Database (gnomAD) for POLR2A, as well as for the genes encoding other pol II subunits. High pLI scores are indicative of intolerance of loss-of-function variation, and Z scores are indicative of intolerance of synonymous or missense variation.29 Additionally, ratios of observed and expected variants were retrieved and acted as a reflection of the degree to which variants within a gene are underreported, indicative of a survival disadvantage.
The CADD score is a tool for scoring the deleteriousness of single-nucleotide variants and insertion and/or deletion variants in the human genome.32 It integrates multiple annotations into one metric by contrasting variants that survived natural selection with simulated variants. CADD scores were obtained for all missense variants in gnomAD, as well as for the missense and nonsense variants of the included individuals. A CADD score >20 suggests pathogenicity because it indicates that the variant is predicted to be among the 1% of most deleterious substitutions that can be done to the human genome.
The distribution of genetic variants throughout POLR2A was assessed on the basis of all POLR2A variants from gnomAD. The interval length (number of amino acid positions) between two sequential missense variants was calculated. A larger interval size signifies a stretch devoid of missense variants and was considered a possible index of pathogenicity for variants found within such an interval. The mean and SD were calculated. The “desert Z score,” indicating to which degree stretches within the gene are devoid of missense variants, was calculated by the formula: (stretch length – mean stretch length)/SD of stretch length.
Structural Evaluation of POLR2A Variants in Pol II
Pol II of S. cerevisiae is extensively studied by X-ray crystallography and cryo-electron microscopy.7, 8, 9, 10, 11, 13, 14, 16, 17, 18, 19, 20 Sequence conservation between human POLR2A and S. cerevisiae rpb1 is high, and thus the available structures can serve as a model for the human protein. To evaluate the putative consequences of POLR2A variants, the structures of RNA polymerase from Protein Data Bank entries PDB: 1i6h, 1r5u, 1r9s, 1twf, 1y1v, 1y1w, 1y1y, 2e2h, 2nvq, 2nvz, 3gtj, 3gtm, 3how, 3hoy, 3po2, 3po3, 4a3l, 4a93, 4bbr, 4gwp, 4v1m, 4v1n, 4v1o, 5sva, 5xog, and 5xon were superimposed by overlaying the coordinates for rpb1 with the algorithm provided by BRAGI.33 Variants were mapped onto the structures and evaluated for their putative impact on local protein folding and enzymatic activity. Figures were prepared in the programs Molscript34 and raster3D.35
Functional Evaluation of Variants in Pol II: POLR2A Variants Expressed in S. cerevisiae
Six POLR2A variants were matched through GeneMatcher at the time the experiment in S. cerevisiae was initiated; therefore six POLR2A variants were functionally evaluated in S. cerevisiae. Plate phenotyping of rpb1 mutants was performed as described before.22 All of the S. cerevisiae strains that we used in this study are listed in Table S1. They were derived from CKY283, CKY718, and CKY721. Shortly, to generate mutant rpb1, point mutations in S. cerevisiae rpb1 were introduced by PCR mutagenesis into the CEN LEU2 plasmid pRS315H3alt-RPB1∗ XmaI 1122-1123 T69 corrected. The CEN LEU2 plasmids, containing mutant rpb1, were transformed into an appropriate Leu− strain with corresponding endogenous rpb1 deletion and complemented with a CEN URA3 WT rpb1 subunit gene expressing wild-type (WT) RPB1. Leu+ transformants were patched on solid medium that lacked leucine and replica-plated to medium that lacked leucine but contained 5-fluoroorotic acid (Thermo Scientific) to select against cells maintaining RPB1 WTURA3 plasmids. Δsub1 or Δdst1 + rpb1 mutant strains for direct testing of double mutant phenotypes were constructed based on CKY721 or CKY718, respectively, and analyzed in the same fashion as rpb1 single mutants.
Assays were performed in biological triplicate in three different backgrounds: WT background, absence of TFIIS (Δdst1 background), and absence of the SUB1 transcription elongation factor (Δsub1 background). Transcriptional activity and genetic interactions were determined. The assays with c.1592A>G (p.Asn531Ser) (p.Asn517Seryeast) and c.2207C>T (p.Thr736Met) (p.Ser713Metyeast) in the WT background were performed in sextuplicate to increase the consistency of the results. p.Ala301Aspyeast was used as a positive control for reduced transcriptional activity,36, 37 and p.Glu1230Lysyeast was used as a positive control for reduced genetic interaction with TFIIS.38
For spot assays, overnight yeast peptone dextrose (YPD) cultures from single colonies grown at 30°C were diluted to an OD600 of 0.15. Five-fold serial dilutions were prepared and spotted on indicated plates and grown for 3 to 6 days at 30°C or 37°C when indicated. YPD medium contained yeast extract (1% w/v final, [Biosciences]), peptone (2% w/v final, [Biosciences]), dextrose (2% w/v final), and bacto agar (2% w/v final, [Biosciences]). Alternative-carbon-source YP media were YP raffinose (2% final w/v) and YP raffinose (2% w/v final) plus galactose (1% w/v final). Synthetic complete medium lacking leucine (SC-Leu) contained: 2gr/l drop-out mix minus leucine (US Biological) and 6.71 gr/l Yeast Nitrogen Base without amino acids and carbohydrate and with ammonium sulfate (YNB) (US Biological) with 2% dextrose. Mycophenolic acid (MPA, [Sigma-Aldrich]) was added to SC-Leu at 20 μg/mL final concentration from a 10 mg/mL stock in ethanol.
Functional Evaluation of Variants in Pol II: POLR2A Variants Expressed in HeLa Cells
Nine POLR2A variants were matched through GeneMatcher at the time the experiment in HeLa cells was initiated, therefore nine POLR2A variants could be functionally evaluated in HeLa cells. The open reading frame of human RPB1 was amplified by PCR with a plasmid expressing RPB1 fusion with a B10 epitope, EGFP, hRPB1, and six His residues36 and introduced into the pDONR201 cloning vector. The POLR2A coding sequence contained a point mutation (AAC>GAC), which resulted in the replacement of asparagine 792 by aspartate and resistance to α-amanitin.39 The observed POLR2A point mutations in the included individuals were introduced through the Quickchange protocol (Stratagene) and verified by DNA sequencing. p.Lys812∗ was used as a representative of c.2098C>T (p.Gln700∗) and c.2203C>T (p.Gln735∗). These two variants could not be designed because amino acid residues Gln700 and Gln735 are localized in front of the built-in resistance to α-amanitin (residue 792). p.Lys812∗ is expected to result in a similar truncated version of the protein as p.Gln700∗ and p.Gln735∗. All RPB1 mutant proteins were tagged by GFP at the N terminus. We created the stable doxycycline inducible cell lines by transfecting pCDNA5/FRQT/TO and pOG44 into HeLa FRT cells carrying the TET repressor by using polyethyleneimine (PEI) and then antibiotic selection. HeLa cells were maintained in DMEM containing 4.5 g/l glucose (GIBCO) supplemented with 10% v/v heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich) and 10 mM L-Glutamine (Sigma-Aldrich) under blasticidin (5 μg/mL) (InvivoGen) and hygromycin B (400 μg/mL) (Roche Diagnostics) selection.
The expression of GFP-tagged RPB1 was induced by treatment with 1 μg/mL doxycycline for 24 h. Cells were spun down for 5 min at 400 g and washed with PBS. Next, cells were lysed in whole-cell lysate (WCL) buffer (50 mM Tris-HCl [pH 8.0], 420 mM NaCl, 10 mM MgCl2, 10% glycerol, 0.1% NP40, 0.5 mM DTT, containing protease inhibitor cocktail), and lysates were subjected to centrifugation at 400 g for 10 min. The supernatant was harvested and stored at −80°C.
For the MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazoliumbromide, Sigma-Aldrich) assay, doxycycline-inducible GFP-RPB1amanitin resistant-expressing mutant HeLa FRT cells were cultured for 3 to 5 days in the presence of 1 μg/mL doxycycline to induce the expression of the mutants. Cells were seeded at 3,000 cells per well in 100 μl normal DMEM in the presence of doxycycline in 96-well plates. After a few hours, the medium was replaced by DMEM with doxycycline, and when indicated, 2.5 μg/mL α-amanitin (Boehringer Mannheim) was added. After 72 h of incubation, MTT was added to a final concentration of 0.5 mg/mL.40 Cells were incubated for 4 h under normal culturing conditions, then the medium was carefully removed, and 100 μL DMSO was added to dissolve the formazan crystals. The absorbance was measured at 570- and 630-nm wavelengths. Relative growth was calculated with the equation (A570,sample – A630,sample)/(A570,untreated – A630,untreated).
GST Pull-Down
A GST-TFIIS recombinant expression construct41 was transformed in BL21DE3; single colonies were picked, grown in LB + ampicillin, and induced for 3 h with 1 mM IPTG when cells reached an OD600 of 0.6. Cells were pelleted by centrifugation and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.0], 300 mM KCl, 2 mM EDTA, 20% sucrose, 0.1% Triton X-100, 0.5 mM PMSF, and 1 mM DTT) containing 50 μL lysozyme (25 mg/mL) and incubated for 10 min on ice. The suspension was freeze-thawed three times and sonicated three times for 20 s. Cells were spun down at 25,000 rpm for 45 min at 4°C, and the supernatant was harvested and stored at −80°C.
Glutathione-agarose (GA) beads (50% slurry) were washed three times with binding buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1 mM ZnCl2, 1 mM DTT, and 0.5 mM PMSF). 20 μL GA beads (Sigma-Aldrich) per reaction were coated with 25 μL GST-TFIIS lysate per reaction for 1 h at 4°C. After this, the GA beads were washed three times with binding buffer and incubated with 300 μg HeLa FRT WCLs (unless otherwise stated) in a final volume of 800 μL (50 mM NaCl at final concentration) for 2 h at 4°C while rotating. Beads were washed three times and proteins were eluted in 15 μL sample buffer by a 5 min incubation at 95°C.
Samples were lysed in sample buffer (160 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, and 0.05% bromophenol blue), run on an 8% SDS-PAGE, and transferred onto a PVDF membrane. The membrane was developed with the appropriate antibodies and ECL. Antibodies came from the following sources: α-Tubulin (DM1A, CP06, Calbiochem) and GFP (#632381, JL-8 Clontech).
Interactome Analysis with Mass Spectrometry
For interactome analysis, HeLa cells were grown, and nuclear extracts were prepared as described.42 The protein concentration was determined with a Bradford assay. GFP affinity purification was essentially performed as described.42 Protein extracts were incubated with GFP-trap beads (Chromotek) or with blocked agarose beads (Chromotek) as a negative control on a rotating wheel at 4°C with 1 mg input protein. Peptides were eluted from the beads by 2 h trypsin incubation in elution buffer (100 mM Tris-HCl [pH 7.5], 2 M urea, and 10 mM DTT). Eluate was collected, and beads were eluted for a second time. Eluates were combined, and trypsin digested overnight. Tryptic digests were desalted with stage tips.43
The samples were cleaned up with in-house-made stage tips. Peptides were separated on a 30-cm pico-tip column (50 μm ID, New Objective) in-house packed with 3 μm aquapur gold C-18 material by applying a gradient (7%–80% ACN 0.1% FA, 140 min), delivered by an easy-nLC 1000 system (LC120, Thermo Scientific), and electro-sprayed directly into an LTQ Orbitrap Mass Spectrometer (Velos, Thermo Scientific). Raw files were analyzed with the MaxQuant software version 1.5.1.0.; oxidation of methionine was set as variable and carbamidomethylation of cysteine was set as fixed modification. The human protein database of UniProt was searched with the peptide and protein false discovery rate set to 1%.
Assessing Phenotype-Genotype Correlation
To assess phenotype-genotype correlation, we evaluated the POLR2A variants for which support for pathogenicity was obtained from positional and functional analyses and the variants for which support for pathogenicity was obtained solely from positional analysis. These variants were considered “probable” to be disease causing, indicating that we are strongly convinced that the individuals’ genotypes are causing the individuals’ phenotypes. We used the phenotypic features of the individuals harboring these variants to delineate the phenotypic spectrum. For the remaining variants, pathogenicity was assessed by determining the degree of overlap. Phenotypic similarity was defined as the presence of at least three phenotypic features of the five most prevalent phenotypic features found in individuals harboring “probable” disease-causing variants in POLR2A. If the overlap in phenotypic features was sufficient, these variants were considered to be “possibly” disease-causing. If the phenotypic overlap was not sufficient or could not be determined, the variant was classified as “unknown” whether this variant is disease-causing or not.
To correlate phenotypic severity to the predicted consequences of the POLR2A variant and to find an explanation for the wide phenotypic spectrum, an ad hoc severity score was conceived.44 It was calculated as follows: each tabulated item that was present (+) in the individual scored 1 point and each absent item (−) scored 0 points (Table 1). The items “sit without support,” “walk well,” and “brain magnetic resonance imaging (MRI) abnormalities” were calculated as follows: “sit without support” at <12 months scored 0 points, at 13–18 months scored 1 point, at 19–30 months scored 2 points, and at >30 months scored 3 points; “walk well” at <24 months scored 0 points, at 25–30 months scored 1 point, at 31–48 months scored 2 points, and at >48 months scored 3 points; no brain abnormalities scored 0 points, wide ventricles and/or delayed myelination scored 1 point, and white matter abnormalities scored 2 points. A severity score of <12 points indicated the phenotype was mild, a score of 12–17 indicated moderate, a score of 18–23 indicated severe, and a score of >23 points indicated profound.
Table 1.
Characteristics of Individuals Harboring a Heterozygous De Novo Variant in POLR2A
| Individual | 5 | 6 | 8 | 12 | 13 | 1 | 16 |
|---|---|---|---|---|---|---|---|
| Variant | p.Gln700∗ | p.Gln735∗ | p.Ser755del | p.Leu1124Pro | p.Lys1125del | p.Pro371Leu | p.Pro1767fs |
| Sex | m | f | m | f | m | f | m |
| Gestation (weeks) | 42 | 28 | Term | 38 | 37 | 41 | 41 |
| Age (years) | 17 | 13 | 3 | 4 | 7 | 7 | 9 |
| General hypotonia | − | + | + | + | + | + | + |
| Strabismus | − | + | + | + | + | + | − |
| Frog position infancy | − | − | + | + | − | − | − |
| Decreased endurance | − | − | + | + | + | − | + |
| Feeding difficulties | − | + | − | + | − | + | + |
| Recurrent RTI | − | − | + | + | − | − | + |
| High forehead | + | + | + | − | + | − | + |
| Disturbed sleeping | + | − | − | − | − | − | − |
| Gastro-esoph. reflux | − | + | − | − | − | − | − |
| High palate | + | − | − | − | + | NA | − |
| Delayed visual matur. | + | − | − | − | − | − | − |
| Microcephaly | − | − | − | − | − | − | − |
| Brachyplagiocephaly | − | − | − | − | + | − | + |
| Muscle atrophy | − | − | − | − | − | − | − |
| Hypertelorism | − | − | − | − | + | + | + |
| Teeth misalignment | − | + | − | + | + | − | + |
| Decreased vision | + | − | − | − | − | + | − |
| Stagnation episodes | − | − | − | − | − | − | − |
| Inguinal hernia | − | − | − | − | − | − | − |
| Decreased fetal movem. | − | − | NA | + | − | − | − |
| Autistic behavior | + | − | − | NA | − | − | − |
| Aggressive behavior | − | − | − | − | − | − | − |
| Failure to thrive | − | − | − | − | − | + | − |
| Pectus excavatum | − | − | − | − | − | − | − |
| Epilepsy | − | − | − | − | − | − | − |
| Sit, no support (months) | 7 | 12 | NA | 12 | 18 | 24 | 24 |
| Walk well (months) | 18 | 30 | >36 | 23 | 24 | 72 | 28 |
| Brain MRI abnormalities | − | Cerebellar atrophy, inferior vermis, small pons, megacisterna magna, and slightly small corpus callosum | Delayed myelination | Delayed myelination | Delayed myelination | − | − |
| Severity score | 6 | 9 | 9 | 9 | 10 | 11 | 11 |
| Severity class | Mild | Mild | Mild | Mild | Mild | Mild | Mild |
| Individual | 4 | 11 | 10 | 15 | 9 | 14 | 2 | 7 | 3 |
|---|---|---|---|---|---|---|---|---|---|
| Variant | p.Tyr669del | p.Tyr1109His | p.Ile848Thr | p.Arg1603His | p.Met769Thr | p.Asn1251Ser | p.Ile457Thr | p.Thr736Met | p.Asn531Ser |
| Sex | f | m | f | f | m | f | m | f | m |
| Gestation | 41 | 40 | 41 | Term | 41 | 40 | 40 | 39 | 30a |
| Age (years) | 11 | 6 | 13 | 7 | 18 | 6 | 4 | 9 | 0 |
| General hypotonia | + | + | + | + | + | + | + | + | NA |
| Strabismus | − | + | − | + | + | + | + | + | NA |
| Frog position infancy | + | + | + | + | + | + | + | + | NA |
| Decreased endurance | + | NA | + | − | + | + | + | + | NA |
| Feeding difficulties | + | − | + | − | + | + | + | + | NA |
| Recurrent RTI∗ | − | + | − | + | − | + | + | + | NA |
| High forehead | − | − | − | − | + | − | + | − | − |
| Disturbed sleeping | + | − | − | + | + | + | + | + | NA |
| Gastro-esoph. reflux | + | − | + | − | − | + | + | + | NA |
| High palate | − | − | − | − | + | − | + | + | Cleft |
| Delayed visual matur. | + | − | − | + | + | − | + | + | NA |
| Microcephaly | − | + | − | + | + | + | − | + | − |
| Brachyplagiocephaly | − | − | + | − | − | + | + | − | − |
| Muscle atrophy | − | − | + | − | + | − | + | + | NA |
| Hypertelorism | − | − | − | − | + | − | + | − | + |
| Teeth misalignment | + | − | − | − | − | − | − | − | − |
| Decreased vision | − | − | − | + | + | − | − | + | NA |
| Stagnation episodes | + | + | − | − | − | − | + | + | NA |
| Inguinal hernia | + | − | − | − | + | − | + | + | NA |
| Decreased fetal movem. | − | − | − | − | − | + | + | − | − |
| Autistic behavior | + | − | − | + | + | − | − | − | NA |
| Aggressive behavior | + | − | + | + | − | + | − | − | NA |
| Failure to thrive | − | − | − | − | − | + | − | + | NA |
| Pectus excavatum | + | − | − | − | + | + | − | − | NA |
| Epilepsy | − | + | − | + | − | − | − | + | NA |
| Sit, no support (months) | 11 | 24 | 24 | Attained | 14 | 48 | 23 | >108 | |
| Walk well (months) | 26 | >72 | >156 | 56 | 27 | 65 | >55 | >108 | |
| Brain MRI abnormalities | − | Wide ventricles, increased T2 signals in nuclei pallidi | Wide ventricles, cystic area in periventricular white matter | Wide ventricles, thin corpus callosum, bilateral loss of white matter | Delayed myelination | Megacisterna magna | Delayed myelination, wide ventricles | Wide ventricles and sulci, cerebellar volume loss, abnormal globus pallidus | Corpus callosum agenesis |
| Severity score | 14 | 14 | 15 | 16 | 19 | 21 | 23 | 25 | NA |
| Severity class | Moderate | Moderate | Moderate | Moderate | Severe | Severe | Severe | Profound | |
Abbreviations are as follows: m = male, f = female, NA = not available, and RTI = respiratory tract infections.
Termination of pregnancy.
Results
Phenotypes of Affected Individuals
Sixteen individuals, all harboring ultra-rare, de novo heterozygous variants in POLR2A, were located via GeneMatcher.30 Other (genetic) causes of the phenotypes of the included individuals were thoroughly excluded. The cohort included three individuals with truncating variants (two stop codons and one frameshift), three individuals with in-frame deletions (IF deletion), and ten individuals with missense variants. Portraits of the individuals are shown in Figure 1A, clinical characteristics of all individuals are summarized in Table 1, and extended individual reports can be found in the Supplemental Data. In summary, eleven individuals were born after a generally uneventful pregnancy and delivery at term and had a normal birth weight and normal perinatal events. Decreased fetal movements were noted in three pregnancies, and one female was born preterm at 28 weeks. One pregnancy carrying a male fetus (individual 3, p.Asn531Ser) was terminated because of corpus callosum agenesis, frontonasal dysplasia, and a cleft lip. The most frequent early and usually striking phenotypic feature was hypotonia. It was noted in fourteen individuals and was profound in nine (Figure S1), evidenced by decreased muscle tone, a frog-like posture in infancy, and reduced spontaneous movements but normal tendon reflexes. Muscles were considered atrophic in four individuals. A muscle biopsy, on suspicion of a myopathy, was performed in four individuals, but the results were inconclusive. In addition, there were symptoms and signs commonly associated with hypotonia; these signs included brachyplagiocephaly (in five individuals), a high arched palate (in five individuals), pectus excavatum (in three individuals), recurrent respiratory tract infections (in eight individuals), and inguinal hernia (in four individuals). The majority of individuals (ten) had feeding difficulties, with gastro-esophageal reflux (in six individuals), resulting in failure to thrive (in three individuals). Ocular signs included strabismus (in eleven individuals) and delayed visual maturation (in six individuals), resulting in decreased vision (in five individuals). Moderate to severe sensorineural hearing loss was reported in two individuals, and conductive hearing loss due to recurrent respiratory tract infections was reported in one individual. Dysmorphic features were generally mild and non-distinctive; these included a high forehead (in seven individuals), hypertelorism (in six individuals), and tooth misalignment (in five individuals), which is defined as an abnormal spacing of the teeth without missing any teeth. Brain MRI performed in eleven individuals revealed white matter abnormalities (in ten individuals), ranging from delayed myelination (in five individuals) to wide lateral ventricles, putatively due to white-matter loss (in four individuals). A thin corpus callosum, a feature consistent with the loss of white matter was noted in two individuals. Additionally, three individuals had cerebellar abnormalities. Severe epilepsy was reported in three individuals. Eight individuals had sleeping difficulties. Autistic (in four individuals) and aggressive (in four individuals) behavior, including extreme behavior such as pica (eating dung), was reported. Hypotonia tended to improve over time, albeit slowly. Later in life, endurance was diminished (in ten individuals). A delayed development involving all domains was noted in all individuals and ranged from mild to severe. The severity of the delay was evaluated by calculating Z scores on the basis of the acquisition of developmental milestones, with higher Z scores indicating later acquisition of milestones. The individuals’ developmental Z scores were stable over time, consistent with a gradual development without developmental catch-up or decline. The Z scores between domains were similar, arguing against developmental-domain-specific delays (Figure 1B). The degree of delay appeared to correlate with the degree of hypotonia. Temporary loss of milestones was reported in four individuals, usually following an infection. Additional metabolic investigations ruled out known inborn errors of metabolism.
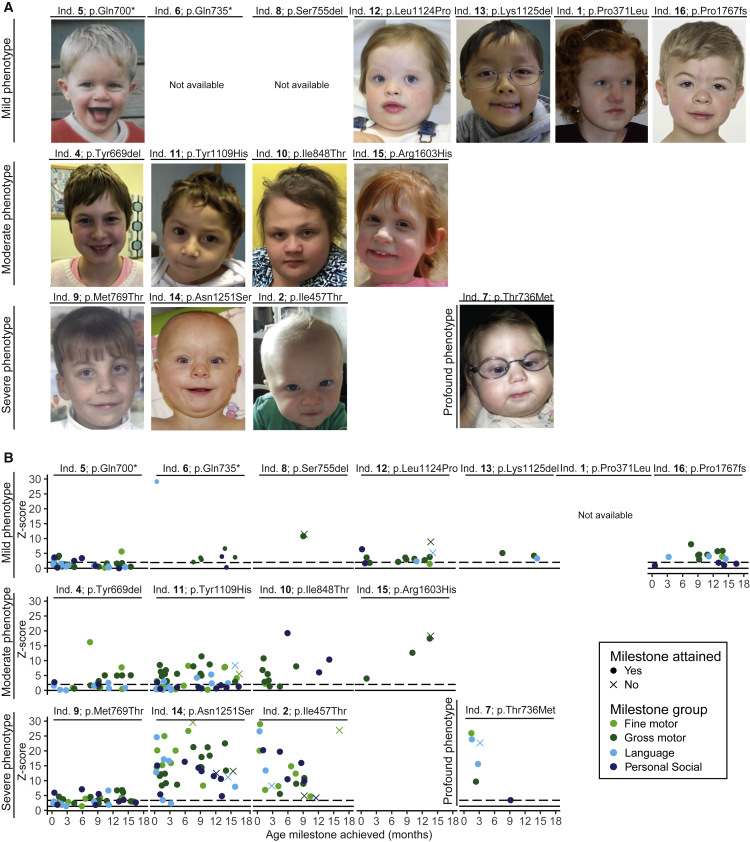
Figure 1.
Photos and Attainment of Developmental Milestones of Individuals with POLR2A Variants
(A) Photos of 13 of the 16 individuals (ind.) harboring a de novo variant in POLR2A, in order of severity score.
(B) Attained and unattained developmental milestones during the first 18 months of life for 14 of the 16 individuals harboring a de novo variant in POLR2A, in order of severity score. For each milestone, Z scores were calculated indicating the delay in attaining that milestone. The higher the Z scores, the greater the developmental delay. The figure demonstrates that in individuals with a mild phenotype, developmental Z scores are mostly <10, in individuals with a moderate phenotype, developmental Z scores are <20, and in individuals with a severe phenotype, developmental Z scores can even reach 30. In addition, among the individuals with a severe phenotype, relatively more (and more early) developmental milestones were not attained by the individual. Individual 3 (p.Asn531Ser) was not included in this figure because the pregnancy was terminated.
Variant Severity Metrics
Large-scale genetic data retrieved from gnomAD from individuals without severe pediatric disease indicate that POLR2A is intolerant of deleterious, heterozygous, protein-changing variants. This is evidenced by a maximal pLI score (1.0) and a very low observed/expected ratio (0.08), indicating intolerance of loss of function variants (Table S2). Moreover, the Z score (7.13) for missense variants is one of the highest of all human protein-coding genes, suggesting that subtle heterozygous changes can also cause a survival disadvantage (Table S2).
An assessment of conservation across species (Figure 2A) revealed that nine out of ten missense variants affect highly conserved amino acid residues (Figure 2B). However, this variant property fails to be discriminatory because the overall degree of conservation of POLR2A is extremely high (Figure 2A). In the pol II core, roughly 50% of residues are identical, and an additional 20% are highly similar between humans and yeast. Similarly, the CADD scores of the individuals’ variants were all above the arbitrary cut-off of 20, indicating that these variants are predicted to be in the 1% of the most deleterious substitutions that can be done to the human genome and thereby suggesting pathogenicity;32 however, most of the CADD scores of the variants observed in the gnomAD cohort of individuals without severe pediatric disease (Figure S2) were also above the cut-off. Thus, these indices of pathogenicity at the amino-acid level lack discriminative power.
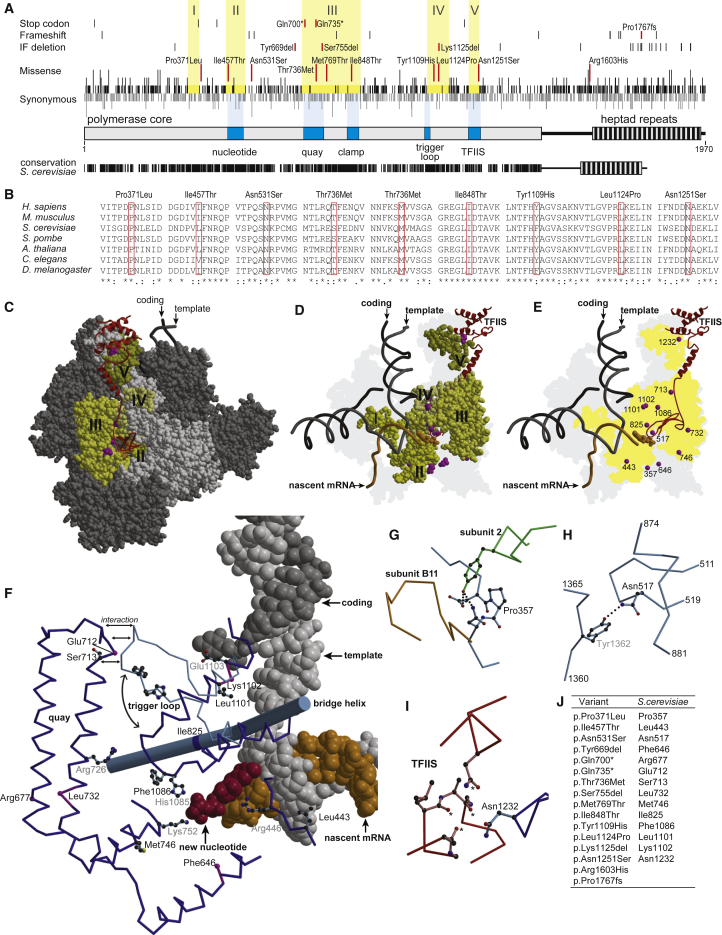
Figure 2.
Structural Evaluation of POLR2A Variants
(A) RPB1 is represented; the core and the C-terminal heptad repeats are shown as bars. Functional elements are highlighted in dark blue and are labeled underneath. The positions of gained stop codons, frameshifts, in-frame deletions (IF deletion), missense mutations, and synonymous mutations at the protein level, as reported in the gnomAD database, are indicated by vertical black strokes. The length of the stroke represents the number of different mutations observed at the DNA level in the corresponding codon. The individuals’ variants are indicated by red strokes and are labeled. Stretches devoid of missense mutations are highlighted by a yellow background and are labeled by Roman numerals. The degree of conservation in humans and S. cerevisiae is depicted underneath, whereby each identical amino acid is represented by a vertical black stroke.
(B) Conservation across species of amino acid residues that are changed in the individuals affected by missense mutations. p.Arg1603His is not conserved and is localized in the region between the pol II core and the heptad repeats, and p.Pro1767fs is localized in the heptad repeat region. For these regions, alignments are not reliable.
(C) A space-filling model of the elongating form of pol II from S. cerevisiae with TFIIS in ribbon representation in red. Subunits other than RPB1 are shown in dark gray. RPB1 is shown in light gray; the positions of individuals’ variants are highlighted in magenta, and stretches devoid of missense variants are shown in shades of yellow labeled by Roman numerals as in (A). The incoming nucleotide is shown as a space-filling model in orange. The template strand, the coding strand, and the nascent mRNA are shown as backbone trace in gray, black, and orange, respectively.
(D) Same as in (C), but omitting subunits other than RPB1. Only the RPB1 stretches devoid of missense variants and the positions of individuals’ variants are shown, and all other parts of RPB1 have been omitted. The 3D-envelope of RPB1 is projected in light gray on the background plane.
(E) Same as in (D), but additionally omitting the RPB1 stretches devoid of missenses variants. The Cα-atoms of amino acid residues affected in individuals’ variants are shown as magenta spheres.
(F) Detailed view at the catalytic center of pol II. The coding strand in dark gray, the template strand in light gray, the nascent mRNA in orange, and the incoming nucleotide in red are shown as a space-filling model. The bridge helix is shown as a rod, and selected other parts of RPB1 as backbone trace. The trigger loop is shown in its open and closed conformation in light and dark blue, respectively. Side chains of amino acid residues affected in individuals with a missense variant and of some residues known to be of catalytic importance are shown in ball-and-stick representation. The Cα-atoms of amino acid residues affected by IF deletions or the gain of a stop codon are shown as spheres in magenta.
(G) The environment of residue Pro357. Elements from RPB1, subunit 2, and the pol II subunit RPB11 are depicted in light blue, green, and orange, respectively. Hydrogen bonds are indicated by dotted lines
(H) The environment of residue Asn517. Elements of RPB1 are shown in blue
(I) The environment of residue Asn1232. Elements of RPB1 and TFIIS are shown in blue and red, respectively. Putative hydrogen bonding partners for Asn1232 in TFIIS are marked by an asterisk.
(J) A table assigning amino acid residues that are affected in individuals to the corresponding residues of RPB1 in S. cerevisiae.
Next, we evaluated whether pathogenicity was related to the variants’ positions within POLR2A. The observation that the individuals’ variants were found throughout the gene argued against a domain-specific deleteriousness. In line with Lelieveld et al. and Havrilla et al.,45, 46 we hypothesized that the distribution of protein-changing POLR2A variants observed in the gnomAD cohort of individuals without severe pediatric disease—apparently tolerated well enough and therefore more likely to be found at positions tolerant to change—could conversely unveil areas intolerant to change. S. cerevisiae tolerates truncations of the CTD region comprising up to 50% of its 26 heptad repeats in rpb1, the ortholog of POLR2A.47, 48 This strongly suggests that the human POLR2A CTD region, consisting of 52 repeats, might be equally tolerant to the loss of a proportion of the heptad repeats. Indeed, the majority of the truncating variants reported in gnomAD—and none of the truncations found in these individuals—are positioned in the distal portion of the CTD region (Figure 2A). In addition, the density of POLR2A missense variants reported in gnomAD is also highest in the CTD region, not only in the distal part but throughout the whole repeat region, indicating that a putative functional loss of a single heptad repeat is tolerated, regardless of its position.
Within the remaining part of POLR2A, missense variants are strikingly unevenly distributed when compared with synonymous variants (Figure 2A). In line with our hypothesis, although the median distance between variants is only two amino acid residues, several stretches, the longest of which spans about 80 amino acid residues, that are devoid of missense variants are observed (Figure 2A). These stretches devoid of missense variants correspond to a number of known functionally important regions of the protein. Nine out of ten missense variants from our individuals, two out of three in-frame deletions, and two out of three truncating variants were positioned within these stretches with desert Z scores that fell between 2.9 and 11.9, indicating intolerance to change in these regions and thereby supporting pathogenicity (Figure 2A and Table 2).
Table 2.
Lines of Evidence for Pathogenicity of Variants in POLR2A
| Individual | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Variant | p.Pro371Leu | p.Ile457Thr | p.Asn531Ser | p.Tyr669del | p.Gln700∗ | p.Gln735∗ | p.Thr736Met | p.Ser755del |
| cDNA change | c.1112C>T | c.1370T>C | c.1592A>G | c.2006_2008delACT | c.2098C>T | c.2203C>T | c.2207C>T | c.2262_2264delCTC |
| Phenotype | ||||||||
| Severity score | 11 | 23 | NA | 14 | 6 | 9 | 25 | 9 |
| Severity class | Mild | Severe | Moderate | Mild | Mild | Profound | Mild | |
| Metrics of Variant Severity | ||||||||
| Yeast conservation | Identical | Similar | Identical | NA | NA | NA | Similar | NA |
| Amino acid stretch | 366–375 | 448–470 | 523–532 | 664–671 | 689–715 | 689–715 | 717–797 | 717–797 |
| Stretch length | 9 | 22 | 9 | 7 | 26 | 26 | 80 | 80 |
| Desert Z score | 0.8 | 2.9 | 0.8 | 0.5 | 3.5 | 3.5 | 11.9 | 11.9 |
| Structural Evaluation | ||||||||
| Position | Funnel | Catalytic site | – | Close to quay | Quay | Quay | Quay | Quay |
| Protein folding | – | – | – | Locally altered | Loss | Loss | – | Locally altered |
| Dominant negative | – | Expected | – | – | – | – | Expected | – |
| Haploinsufficiency | – | – | – | Putative | Expected | Expected | – | Putative |
| Functional Evaluation | ||||||||
| Yeast mutation | NA | Leu443Thr | Asn517Ser | NA | NA | NA | Ser713Met | Leu732del |
| Yeast growth (WT) | NA | Normal | Normal | NA | NA | NA | Normal | Normal |
| Yeast growth (Δdst1; Δsub1) | NA | Aberrant | Normal | NA | NA | NA | Aberrant | Aberrant |
| Yeast galactose sensitivity | NA | Yes | No | NA | NA | NA | Yes | Yes |
| Yeast MPA sensitivity | NA | Yes | No | NA | NA | NA | Yes | Yes |
| HeLa cell viability | NA | = | = | NA | ↓↓↓ | ↓↓↓ | ↓↓↓ | ↓↓↓ |
| Disease-causing variant | Possible | Probable | Unknown | Possible | Probable | Probable | Probable | Probable |
| Individual | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|
| Variant | p.Met769Thr | p.Ile848Thr | p.Tyr1109His | p.Leu1124Pro | p.Lys1125del | p.Asn1251Ser | p.Arg1603His | p.Pro1767fs |
| Mutation | c.2306T>C | c.2543T>C | c.3325T>C | c.3371T>C | c.3373_3375delAAG | c.3752A>G | c.4808G>A | c.5298dup |
| Phenotype | ||||||||
| Severity score | 19 | 15 | 14 | 9 | 10 | 21 | 16 | 11 |
| Severity class | Severe | Moderate | Moderate | Mild | Mild | Severe | Moderate | Mild |
| Metrics of Variant Severity | ||||||||
| Yeast conservation | Identical | Identical | Similar | Identical | NA | Identical | Other | NA |
| Amino acid stretch | 717–797 | 836–877 | 1086–1135 | 1086–1135 | 1086–1135 | 1218–1254 | 1603–1604 | 1766–1769 |
| Stretch length | 80 | 41 | 49 | 49 | 49 | 36 | 1 | 3 |
| Desert Z score | 11.9 | 5.8 | 7.1 | 7.1 | 7.1 | 5.0 | −0.4 | −0.1 |
| Structural Evaluation | ||||||||
| Position | Catalytic site | Bridge helix | Trigger loop | Trigger loop | Trigger loop | TFIIS interact. | – | Hepta repeats |
| Protein folding | – | – | – | – | Locally altered | – | – | Core normal |
| Dominant negative | Expected | – | Expected | Expected | – | – | – | – |
| Haploinsufficiency | – | – | – | – | Putative | – | – | – |
| Functional Evaluation | ||||||||
| Yeast mutation | NA | NA | NA | Leu1101Pro | NA | Asn1232Ser | NA | NA |
| Yeast growth (WT) | NA | NA | NA | Aberrant | NA | Normal | NA | NA |
| Yeast growth (Δdst1; Δsub1) | NA | NA | NA | Aberrant | NA | Normal | NA | NA |
| Yeast galactose sensitivity | NA | NA | NA | Yes | NA | No | NA | NA |
| Yeast MPA sensitivity | NA | NA | NA | Yes | NA | No | NA | NA |
| HeLa cell viability | NA | NA | NA | ↓↓↓ | NA | = | = | NA |
| Disease-causing variant | Probable | Possible | Probable | Probable | Probable | Probable | Possible | Probable |
Abbreviations are as follows: NA = not available, WT = wildtype, and MPA = mycophenolic acid. A dash indicates results were unknown.
Structural Evaluation of POLR2A Variants in Pol II
Most of the amino residues affected in the individuals have identical or similar residues in the S. cerevisiae counterpart (Figures 2A, 2B, and 2J), for which high-resolution structural information is available. This allows for structural evaluation of the variants in pol II. Variants p.Gln700∗ and p.Gln735∗ result in a truncation that leads to missing more than half of the protein. If translation is not already prevented by nonsense-mediated decay, the truncated protein is still not expected to form a stable fold with the second largest and core subunit of pol II, RPB2, and it lacks binding sites for several other subunits (Table 2). It is therefore most likely that effects observed with p.Gln700∗ and p.Gln735∗ result from RPB1 haploinsufficiency. In support of this, mass spectrometry analyses showed that the variant p.Lys812∗ (which we used as a representative of p.Gln700∗ and p.Gln735∗ because p.Lys812∗ is similarly truncated, see Material and Methods) could only associate with RPABC3 (POLR2H) but not with any of the other pol II subunits (Figure 3C). Variant c.5298dup (p.Pro1767fs) is located in the proximal half of the CTD and results in the loss of the last 200 amino acids, corresponding to about 50% of the heptad repeats. The folding of the polymerase core is not expected to be affected by this truncation.49, 50 Therefore, p.Pro1767fs is expected to form pol II, but because the CTD region of RPB1 is too short, pol II function might be affected.
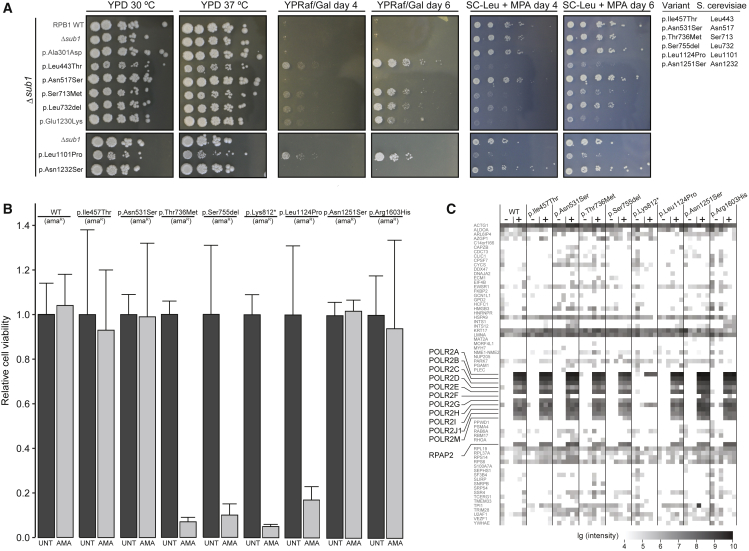
Figure 3.
Functional Evaluation of POLR2A Variants
(A) A growth assay in S. cerevisiae of six rpb1 mutants: p.Ile457Thr (p.Leu443Thr), p.Asn531Ser (p.Asn517Ser), p.Thr736Met (p.Ser713Met), p.Ser755del (p.Leu732del), p.Leu1124Pro (p.Leu1101Pro), and p.Asn1251Ser (p.Asn1232Ser) are compared to wild type (WT). p.Ala301Asp demonstrates a positive control for reduced transcriptional activity, and p.Glu1230Lys demonstrates a positive control for reduced genetic interaction with TFIIS. YPD 30°C depicts growth under normal circumstances, and YPD 37°C depicts growth under stress induced by increased temperature. YPRaf/Gal day 4 and day 6 depict growth under stress induced by increased inappropriate GAL 3′ termination on day 4 and day 6 of culture, respectively. SC-LEU + MPA day 4 and day 6 depict growth under stress induced by the lack of leucine and the addition of mycophenolic acid (MPA) on day 4 and day 6 of culture, respectively.
(B) HeLa cell viability. The individuals’ variants were introduced in an α-amanitin-resistant version of POLR2A (amaR) and expressed from a doxycycline-inducible promoter as a GFP fusion construct in HeLa cells. After the induction of expression, endogenous pol II was left untreated (dark gray bars) or inhibited by the application of α-amanitin (light gray bars), and cell viability was determined. Error bars represent the standard deviation. p.Lys812∗ was used as a representative of p.Gln700∗ and p.Gln735∗ because it is similarly truncated (see Material and Methods).
(C) Interactome analysis of wild-type (WT) and mutant versions of RPB1 using mass spectrometry. Nuclear extracts from HeLa cells expressing GFP-tagged WT or mutant versions of RPB1 were used for precipitations with control agarose beads (−) or GFP-affinity beads (+) in triplicate. In total, 1,133 proteins were identified by more than two peptides via intensity-based absolute quantification (IBAQ) mass spectroscopy. Known members of pol II (highlighted) and a random selection of 5% of the other proteins are shown. The summed peptide intensities of identified proteins were logarithmically transformed to the basis of ten and are represented by shades of gray. Note, intensities smaller than 104 are not obtained due to technical constrains. p.Lys812∗ was used as a representative of p.Gln700∗ and p.Gln735∗.
The IF deletions c.2006_2008delACT (p.Tyr669del) (p.Phe646yeast) and c.2262_2264delCTC (p.Ser755del) (p.Leu732yeast) are localized in close proximity to the quay, whereas the IF deletion c.3373_3375delAAG (p.Lys1125del) (p.Lys1102yeast) is localized in the trigger loop (Figures 2C–2F). The deletions are predicted to alter the local protein fold and to have a strong impact on catalytic activity. It is difficult to predict to what extent the overall protein fold and the ability to assemble into pol II are impaired, but such impairment would shift the properties from severe dominant-negative to mild haploinsufficiency. In case of p.Ser755del, mass spectrometry data showed that the interaction with other pol II subunits is largely unaltered, showing that formation of the pol II complex is not affected (Figure 3C).
Interestingly, most missense variants are centered around the catalytic site (Figures 2C–2F and Table 2): c.3325T>C (p.Tyr1109His) (p.Phe1086yeast) and c.3371T>C (p.Leu1124Pro) (p.Leu1101yeast) are positioned in the trigger loop, p.Thr736Met (p.Ser713yeast) in the opposing quay, c.1370T>C (p.Ile457Thr) (p.Leu443yeast) and c.2306T>C (p.Met769Thr) (p.Met746yeast) in the nucleotide binding site, c.1112C>T (p.Pro371Leu) (p.Pro357yeast) in the funnel (Figure 2G), and c.2543T>C (p.Ile848Thr) (p.Ile825yeast) in the bridge helix (Figures 2C–2F and Table 2). These POLR2A variants are likely to reduce the transcriptional pausing, elongation, and/or back-tracking activities of pol II without affecting transcription initiation. Indeed, all tested missense RPB1 variants were found to interact with other pol II subunits, indicating that complex formation is largely unaltered (Figure 3C).
The missense mutation c.3752A>G (p.Asn1251Ser) (p.Asn1232yeast) is located distantly from the catalytic site, but it is localized in the interaction surface for transcription-elongation factor TFIIS, which is required for back-tracking of pol II due to elongation blocks or nucleotide mis-incorporation (Figure 2I). TFIIS interaction mutants of pol II would have both an increased error rate and reduced elongation rates. The consequences of the missense variants p.Asn531Ser (p.Asn517yeast) (Figure 2H) and c.4808G>A (p.Arg1603His) are more difficult to predict. p.Asn531 is not directly associated with a structural element involved in catalysis (Figure 2H). p.Arg1603His is localized in the region that connects the RPB1 core to the heptad repeats (Figure 2A). No structural information is available for this region, and it is only partially conserved between human and S. cerevisiae RPB1 (Figure 2A).
Altogether, structural evaluation of POLR2A variants resulted in the expected haploinsufficiency for p.Gln700∗ and p.Gln735∗, putative haploinsufficiency for the IF deletions, and an expected dominant-negative effect for the missense variants p.Ile457Thr, p.Thr736Met, p.Met769Thr, p.Tyr1109His, and p.Leu1124Pro (Table 2).
Functional Evaluation of POLR2A Variants in Pol II
The consequences of POLR2A variants on pol II function were investigated in S. cerevisiae and HeLa cells for six and nine of the individuals’ variants, respectively, representing the whole clinical spectrum (Table 2).
In S. cerevisiae, in a WT genetic background, normal growth was observed in all but one variant (p.Leu1124Pro [p.Leu1101Proyeast]) under optimal conditions, as well as under temperature stress conditions (YPD at 37°C) (Table 2 and Figure S3). In genetic backgrounds lacking transcription factors Dst1 (yeast ortholog of TFIIS) or Sub1 (yeast ortholog of PC4), aberrant growth surfaced in four out of six variants (p.Leu1124Pro [p.Leu1101Proyeast], p.Ile457Thr [p.Leu443Thryeast], p.Thr736Met [p.Ser713Metyeast], and p.Ser755del [p.Leu732delyeast]) when compared with WT and the positive controls for Δdst1 (p.Glu1230Lysyeast) (Table 2 and Figure S3) and Δsub1 (p.Ala301Aspyeast) (Figure 3A and Table 2). A read-through transcription assay, in which GAL7 production is inhibited in WT (gal10Δ56) cells, causing galactose sensitivity on YPRaf/Gal,23 also shows aberrant growth for these four variants (Figures 3A and S3 and Table 2). Although milder, these four variants also exhibited aberrant growth when exposed to the nucleotide synthesis inhibitor mycophenolic acid (MPA) in one or both of these genetic backgrounds (Figures 3A and S3 and Table 2). Disturbances in both genetic backgrounds (Δdst1 and Δsub1) are suggestive of reduced transcriptional fidelity.
Cell viability was assessed with an MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazoliumbromide) assay in doxycycline-inducible GFP-RPB1amanitin resistant-expressing mutant HeLa FRT cells, in which the endogenous WT RPB1 was annihilated with the pol-II-specific drug α-amanitin. Cell viability was grossly reduced in mutants expressing variants p.Thr736Met, p.Ser755del, p.Lys812∗, and p.Leu1124Pro (Figure 3B), supporting the pathogenicity of these variants. No significant effects were seen for the variants p.Ile457Thr, p.Asn531Ser, p.Asn1251Ser, and p.Arg1603His (Figure 3B). For all mutants, the amounts of RPB1 protein were similar to the protein amount of WT RPB1 (Figure S4). Binding affinity toward TFIIS, a key transcription factor, was unaffected for all mutants. This included the p.Asn1251Ser mutant, despite its location in the TFIIS binding site (Figure S4). Taken together, the characterization in S. cerevisiae and MTT cell viability in HeLa cells provide evidence for pathogenicity of variants located close to the catalytic core, but not for p.Asn531Ser, p.Asn1251Ser, and p.Arg1603His (Table 2).
Assessing Phenotype-Genotype Correlation
To delineate the phenotypic spectrum that could be attributed to POLR2A variants, we focused on variants considered to be probable to cause disease. Positional and functional evaluation of the POLR2A variants’ effects on pol II function supported pathogenicity of the missense variants p.Ile457Thr, p.Thr736Met, and p.Leu1124Pro, as well as of the IF deletion p.Ser755del and the truncated variants p.Gln700∗ and p.Gln735∗ (Table 2), and solely positional evaluation supported pathogenicity for the missense variants p.Met769Thr, p.Ile848Thr, p.Tyr1109His, and p.Asn1251Ser and for the IF deletion p.Lys1125del (Table 2). The phenotypic features of the eleven individuals harboring these variants were used to delineate the POLR2A phenotypic spectrum (Table 2). These phenotypic features included profound general hypotonia (in ten individuals), as evidenced by a frog position in infancy (in eight individuals), strabismus (in nine individuals), decreased endurance (in eight individuals), and feeding difficulties (in seven individuals) (Table 1). The pathogenicity of the remaining variants was assessed by determining the degree of phenotypic overlap. Individual 1 (p.Pro371Leu), individual 4 (p.Tyr669del), individual 15 (p.Arg1603His), and individual 16 (p.Pro1767fs) presented with clinical phenotypes clearly fitting the phenotypes of the other POLR2A individuals because all had at least three of the five most prevalent POLR2A symptoms (Table 1), and therefore these variants were considered to be possibly disease-causing (Table 2). For individual 3 (p.Asn531Ser), the phenotypic overlap could not be assessed because the pregnancy had been terminated, and it is thus unknown whether this variant is disease-causing (Table 2).
We noted that the degree of developmental delay correlated with the predicted consequences of the variant: in the individuals with variants predicted to cause haploinsufficiency, we noted relatively low Z scores (mean Z score 1.74 ± 3.13), but variants expected to cause a dominant-negative effect were associated with appreciably higher Z scores (median Z score 4.23 ± 3.96, p < 0.0001) and thus with more severe delays (Figure 1B). To assess this further, we calculated a severity score that summarizes the severity of the clinical phenotype for POLR2A. The severity scores of individuals harboring variants with expected or putative haploinsufficiency (mean severity score 10, range 6–14 points) were lower than in individuals harboring variants with expected dominant-negative effects (mean severity score 17, range 6–25 points, Table 1). This suggests that a missense mutation exerting a dominant-negative effect is more likely to result in a severe phenotype (severity score >15) than a mutation inducing haploinsufficiency.
Discussion
In this study, we applied an iterative approach to assess the pathogenicity of identified variants in POLR2A. The main argument supporting the pathogenicity of the identified variants is that all variants occurred de novo.27 In addition, POLR2A is, overall, very intolerant of deleterious, heterozygous, protein-changing variants. Furthermore, nine out of ten missense variants affected highly conserved amino acid residues that are localized at important functional regions of the gene. Functional analyses support pathogenicity of the variants p.Ile457Thr, p.Thr736Met, p.Gln700∗, p.Gln735∗, p.Ser755del, and p.Leu1124Pro, although we note that both yeast and HeLa cells are model systems that might not fully recapitulate the complex developmental disease of the individuals reported here. Although we did not demonstrate support from functional analyses for the variants p.Asn531Ser and p.Asn1251Ser, the pathogenicity of p.Asn1251Ser is strongly supported by the conservation of the amino acid residue and the importance of the region for interaction with TFIIS. Altogether, on the basis of the results of both predictive and functional analyses, eleven variants were classified as probably disease-causing (Table 2).
Deep phenotyping, including quantification of phenotypic severity of the individuals, allowed us to delineate the phenotypic spectrum that could be ascribed to the pathogenic POLR2A variants. The severe end of the spectrum is characterized by profound infantile-onset hypotonia and developmental delay, and is further accompanied by strabismus, decreased endurance, and feeding difficulties. Determining the degree of clinical overlap between the individuals harboring probably disease-causing variants and the remaining individuals allowed us to classify four additional variants as possibly disease-causing. Additional support for the pathogenicity of the variant p.Asn531Ser, other than its de novo occurrence, conservation across species, and a severe phenotype, could not be determined, so this variant was classified as of unknown significance.
Thus, the iterative process described here allowed us to delineate pathogenicity for the majority—but not all—of the individuals’ variants. Given the current knowledge, evidence that either confirms or excludes pathogenicity for four possibly disease-causing variants and the one variant of unknown significance will be hard to obtain. This is illustrated by the variant p.Arg1603His. Support for its pathogenicity is obtained from its de novo occurrence and from the phenotypic overlap of individual 15 with individuals harboring probably disease-causing variants in POLR2A; this overlap includes symptoms such as extreme hypotonia with a frog-like positioning of the legs, developmental delay, strabismus, recurrent respiratory tract infections, and wide ventricles, a thin corpus callosum, and bilateral white-matter loss on brain MRI. However, arguments against its pathogenicity are that the variant has been reported three times in gnomAD and that it is part of a poorly conserved region with no known functional importance. We stress the importance of including these unresolved issues when describing novel disease-causing genes because this aids the subsequent resolution of variant pathogenicity when more individuals are described.
In our search for lines of evidence to support pathogenicity, we found an extra line of evidence to support pathogenicity of the variants in POLR2A in the design of a desert Z score. While the CADD score,32 which is usually seen as an estimate of the likelihood of a variant being pathogenic, did not differentiate between variants reported in gnomAD and the missense variants reported here, the variants that we report appeared to cluster within POLR2A in large regions devoid of apparently harmless variants reported in gnomAD. We calculated the desert Z score to reflect this property for all variants within POLR2A. This score confirmed not only that the size of these regions exceeded the size that would be expected by chance (the largest region had a Z score of 11.9), but also confirmed the clustering of individuals’ variants within these regions. The relevance of this finding was supported by subsequent structural analyses that unveiled the functional importance of these “desert regions.” We therefore anticipate, in line with Lelieveld et al. and Havrilla et al.,45, 46 that this property might be helpful to identify within-gene regions that are relatively intolerant of genetic variants and are likely to be of functional importance.
The quantification of phenotypic severity unveiled that individuals harboring heterozygous truncating variants that are incapable of proper pol II formation presented with a mild phenotype, whereas individuals harboring heterozygous missense variants that allow the formation of pol II exhibited the most severe phenotype. This observation implies that the presence of a malfunctioning species of pol II is more detrimental than the reduced availability of pol II alone. We propose that the aberrant pol II enzymes are capable of the proper assembly of the pol II machinery at the transcription start sites, but subsequent elongation of the nascent RNA occurs at reduced rates and possibly with an increased error rate, blocking access to and progression of WT pol II on the same DNA strand. Altogether, this could greatly influence the dynamics of pol II initiation and release, both of which are essential components of transcriptional regulation.21 The results reported here direct further mechanistic studies to investigate this defective elongation hypothesis.
Malfunctioning pol II has deleterious consequences on transcription. To date, none of the eleven other pol II subunits have been implicated in human disease. Interestingly, we noticed some phenotypic overlap between individuals harboring variants in POLR2A and individuals harboring variants in subunits of pol III. Mutations in POLR1C (MIM: 610060, encoding a subunit that is part of both pol III and pol I), POLR3A (MIM: 614258), and POLR3B (MIM: 614366) have been described to cause hypomyelinating leukodystrophy 11 (HLD11, MIM: 616494), 7 (HLD7, MIM: 607694), and 8 (HLD8, MIM: 614381), respectively. These three diseases are summarized under the name POLR3-related leukodystrophy.51 Recently, two individuals with HLD were reported to harbor mutations in POLR3K,52 thereby expanding the list of potential genetic defects underlying POLR3-related leukodystrophy. In addition to hypomyelination on brain imaging, the clinical phenotype of POLR3-related leukodystrophy is characterized by progressive cerebellar dysfunction and cognitive dysfunction. Non-neurological features are abnormal dentition and hypogonadotropic hypogonadism. Phenotypic features of POLR3-related leukodystrophy that are also noted in POLR2A individuals are delayed myelination (5/15), white-matter loss (6/15), and tooth misalignment (5/15). However, the cellular functions of the three nuclear RNA polymerases are different; pol III and pol II are responsible for tRNA and mRNA synthesis, respectively. This can be reconciled by the proposal that mutations in both pol II and pol III subunits can affect protein synthesis via different mechanisms, and this can result in similar clinical phenotypes.
In addition to POLR3-related leukodystrophy, POLR1A has been associated with both acrofacial dysostosis53 (MIM: 616462) and severe neurodegenerative disease with ataxia, psychomotor retardation, cerebellar and cerebral atrophy, and leukodystrophy;54 POLR3A is also, in addition to HLD7, associated with Wiedemann-Rautenstrauch syndrome, a neonatal progeroid syndrome55 (MIM: 264090), and has been discussed as potential cause of hereditary ataxia and spastic paraparesis,56, 57 implying a broader phenotypic range for POLR3A mutations.
In summary, we here report that heterozygous de novo variants in POLR2A, which is indispensable for the synthesis of mRNA and several non-coding RNAs as it encodes the RPB1 subunit of pol II, can result in a neurodevelopmental syndrome characterized by profound infantile-onset hypotonia and developmental delay. We conclude that the clinical consequences of probably disease-causing variants in POLR2A are dependent on their effect on pol-II-mediated transcription because POLR2A variants predicted to result in loss of RPB1 protein are better tolerated than missense variants, which we propose can cause a dominant-negative effect on pol-II-dependent transcription.
Acknowledgements
We thank all included individuals and their families for their cooperation. We thank Ton A.J. van Essen, who unfortunately passed away during the completion of this manuscript. We are indebted to Craig Kaplan for providing yeast strains, plasmids, and insightful discussions. The human RPB1 cDNA was kindly provided by P. Cook. The study was supported by the following: grant 17-29423A from the Czech Ministry of Health for individuals with the p.Pro371Leu and p.Met769Thr variants; grant R6-388/WT100127 from the Oxford NIHR (National Institute for Health Research) Biomedical Research Centre and the Health Innovation Challenge Fund (a parallel funding partnership between the Wellcome Trust and the Department of Health) for the individual with the p.Gln735∗ variant; funding from the Duke Genome Sequencing Clinic, supported by the Duke University Health System, for the individual with the p.Ile848Thr variant; grant GSP15001 from the Telethon Foundation Telethon Undiagnosed Diseases Program for the individual with the p.Tyr1109His variant; and funding from Mining for Miracles (BC Children’s Hospital Foundation) and Genome British Columbia through the CAUSES (Clinical Assessment of the Utility of Sequencing as a Service) study for the individual with the p.Lys1125del variant. The investigators include Shelin Adam, Christèle du Souich, Alison M. Elliott, Anna Lehman, Jill Mwenifumbo, Tanya N. Nelson, Clara van Karnebeek, and Jan M. Friedman, and bioinformatics support was provided by the lab of Wyeth Wasserman. The work of H.T.M.T. is supported by the SFB850 and SFB992 programs of the Deutsche Forschungsgesellschaft (DFG). The mass spectrometry analysis was supported by the “Proteins at Work” program of the Netherlands Organization for Scientific Research (NWO, 184.032.201). The work of H.A. Haijes is supported by the personal Alexandre Suerman Stipend of the University Medical Centre Utrecht.
Published: July 25, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.06.016.
Web Resources
GeneMatcher, https://www.genematcher.org
OMIM, https://www.omim.org
Protein Data Bank, http://www.rcsb.org
UniProt, https://www.uniprot.org
Declaration of Interests
S.P. discloses that he is an employee of AstraZeneca. The other authors declare no competing interests.
Supplemental Data
References
- 1.Wintzerith M., Acker J., Vicaire S., Vigneron M., Kedinger C. Complete sequence of the human RNA polymerase II largest subunit. Nucleic Acids Res. 1992;20:910. doi: 10.1093/nar/20.4.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mita K., Tsuji H., Morimyo M., Takahashi E., Nenoi M., Ichimura S., Yamauchi M., Hongo E., Hayashi A. The human gene encoding the largest subunit of RNA polymerase II. Gene. 1995;159:285–286. doi: 10.1016/0378-1119(95)00081-g. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg R.D. Eukaryotic transcriptional control. Trends Cell Biol. 1999;9:M46–M49. [PubMed] [Google Scholar]
- 4.Roeder R.G., Rutter W.J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 5.Thomas M.C., Chiang C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 6.Sainsbury S., Bernecky C., Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:129–143. doi: 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- 7.Bushnell D.A., Westover K.D., Davis R.E., Kornberg R.D. Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 8.Sainsbury S., Niesser J., Cramer P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2013;493:437–440. doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- 9.Plaschka C., Larivière L., Wenzeck L., Seizl M., Hemann M., Tegunov D., Petrotchenko E.V., Borchers C.H., Baumeister W., Herzog F. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- 10.Robinson P.J., Trnka M.J., Bushnell D.A., Davis R.E., Mattei P.J., Burlingame A.L., Kornberg R.D. Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell. 2016;166:1411–1422.e16. doi: 10.1016/j.cell.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westover K.D., Bushnell D.A., Kornberg R.D. Structural basis of transcription: Nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Cramer P. RNA polymerase II structure: From core to functional complexes. Curr. Opin. Genet. Dev. 2004;14:218–226. doi: 10.1016/j.gde.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Cheung A.C., Sainsbury S., Cramer P. Structural basis of initial RNA polymerase II transcription. EMBO J. 2011;30:4755–4763. doi: 10.1038/emboj.2011.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gnatt A.L., Cramer P., Fu J., Bushnell D.A., Kornberg R.D. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 15.Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehara H., Yokoyama T., Shigematsu H., Yokoyama S., Shirouzu M., Sekine S.I. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science. 2017;357:921–924. doi: 10.1126/science.aan8552. [DOI] [PubMed] [Google Scholar]
- 17.Kettenberger H., Armache K.J., Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 18.Sydow J.F., Brueckner F., Cheung A.C., Damsma G.E., Dengl S., Lehmann E., Vassylyev D., Cramer P. Structural basis of transcription: Mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol. Cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Cheung A.C., Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 20.Walmacq C., Cheung A.C., Kireeva M.L., Lubkowska L., Ye C., Gotte D., Strathern J.N., Carell T., Cramer P., Kashlev M. Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol. Cell. 2012;46:18–29. doi: 10.1016/j.molcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steurer B., Janssens R.C., Geverts B., Geijer M.E., Wienholz F., Theil A.F., Chang J., Dealy S., Pothof J., van Cappellen W.A. Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II. Proc. Natl. Acad. Sci. USA. 2018;115:E4368–E4376. doi: 10.1073/pnas.1717920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan C.D., Jin H., Zhang I.L., Belyanin A. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timmers H.T.M., Tora L. Transcription buffering: A balancing act between mRNA synthesis and mRNA degradation. Mol. Cell. 2018;72:10–17. doi: 10.1016/j.molcel.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 25.Sugaya K. Amino acid substitution of the largest subunit of yeast RNA polymerase II: Effect of a temperature-sensitive mutation related to G1 cell cycle arrest. Curr. Microbiol. 2003;47:159–162. doi: 10.1007/s00284-002-3959-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q.Q., Li F., Fu Z.Y., Liu X.B., Yuan K., Fang Y., Liu Y., Li G., Zhang X.S., Chong K. Intact Arabidopsis RPB1 functions in stem cell niches maintenance and cell cycling control. Plant J. 2018;95:150–167. doi: 10.1111/tpj.13939. [DOI] [PubMed] [Google Scholar]
- 27.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 29.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray O.P. The Denver scale. Dev. Med. Child Neurol. 1972;14:666–667. doi: 10.1111/j.1469-8749.1972.tb02654.x. [DOI] [PubMed] [Google Scholar]
- 32.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schomburg D., Reichelt J. BRAGI – A comprehensive protein modeling program system. J. Mol. Graph. 1988;6:161–165. [Google Scholar]
- 34.Kraulis P.J. Molscript – A program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 35.Merritt E.A., Murphy M.E.P. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 36.Sugaya K., Vigneron M., Cook P.R. Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J. Cell Sci. 2000;113:2679–2683. doi: 10.1242/jcs.113.15.2679. [DOI] [PubMed] [Google Scholar]
- 37.Sugaya K., Sasanuma S., Cook P.R., Mita K. A mutation in the largest (catalytic) subunit of RNA polymerase II and its relation to the arrest of the cell cycle in G(1) phase. Gene. 2001;274:77–81. doi: 10.1016/s0378-1119(01)00615-1. [DOI] [PubMed] [Google Scholar]
- 38.Malagon F., Kireeva M.L., Shafer B.K., Lubkowska L., Kashlev M., Strathern J.N. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics. 2006;172:2201–2209. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen V.T., Giannoni F., Dubois M.F., Seo S.J., Vigneron M., Kédinger C., Bensaude O. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 1996;24:2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baas R., Lelieveld D., van Teeffelen H., Lijnzaad P., Castelijns B., van Schaik F.M., Vermeulen M., Egan D.A., Timmers H.T., de Graaf P. A novel microscopy-based high-throughput screening method to identify proteins that regulate global histone modification levels. J. Biomol. Screen. 2014;19:287–296. doi: 10.1177/1087057113515024. [DOI] [PubMed] [Google Scholar]
- 41.Pereira L.A., van der Knaap J.A., van den Boom V., van den Heuvel F.A., Timmers H.T. TAF(II)170 interacts with the concave surface of TATA-binding protein to inhibit its DNA binding activity. Mol. Cell. Biol. 2001;21:7523–7534. doi: 10.1128/MCB.21.21.7523-7534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baymaz H.I., Spruijt C.G., Vermeulen M. Identifying nuclear protein-protein interactions using GFP affinity purification and SILAC-based quantitative mass spectrometry. Methods Mol. Biol. 2014;1188:207–226. doi: 10.1007/978-1-4939-1142-4_15. [DOI] [PubMed] [Google Scholar]
- 43.Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 44.Haijes H.A., Jaeken J., Foulquier F., van Hasselt P.M. Hypothesis: Lobe A (COG1-4)-CDG causes a more severe phenotype than lobe B (COG5-8)-CDG. J. Med. Genet. 2018;55:137–142. doi: 10.1136/jmedgenet-2017-104586. [DOI] [PubMed] [Google Scholar]
- 45.Lelieveld S.H., Wiel L., Venselaar H., Pfundt R., Vriend G., Veltman J.A., Brunner H.G., Vissers L.E.L.M., Gilissen C. Spatial clustering of de novo missense mutations identifies candidate neurodevelopmental disorder-associated genes. Am. J. Hum. Genet. 2017;101:478–484. doi: 10.1016/j.ajhg.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Havrilla J.M., Pedersen B.S., Layer R.M., Quinlan A.R. A map of constrained coding regions in the human genome. Nat. Genet. 2019;51:88–95. doi: 10.1038/s41588-018-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scafe C., Chao D., Lopes J., Hirsch J.P., Henry S., Young R.A. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 48.Eick D., Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 49.Nonet M., Sweetser D., Young R.A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- 50.Rosonina E., Blencowe B.J. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA. 2004;10:581–589. doi: 10.1261/rna.5207204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernard G., Vanderver A. POLR3-related leukodystrophy. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. GeneReviews. 2012. Updated: May 11, 2017. [Google Scholar]
- 52.Dorboz I., Dumay-Odelot H., Boussaid K., Bouyacoub Y., Barreau P., Samaan S., Jmel H., Eymard-Pierre E., Cances C., Bar C. Mutation in POLR3K causes hypomyelinating leukodystrophy and abnormal ribosomal RNA regulation. Neurol Genet. 2018;4:e289. doi: 10.1212/NXG.0000000000000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weaver K.N., Watt K.E.N., Hufnagel R.B., Navajas Acedo J., Linscott L.L., Sund K.L., Bender P.L., König R., Lourenco C.M., Hehr U. Acrofacial dysostosis, Cincinatti type, a mandibulofacial dysostosis syndrome with limb anomalies, is caused by POLR1A dysfunction. Am. J. Hum. Genet. 2015;96:765–774. doi: 10.1016/j.ajhg.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kara B., Köroğlu Ç., Peltonen K., Steinberg R.C., Maraş Genç H., Hölttä-Vuori M., Güven A., Kanerva K., Kotil T., Solakoğlu S. Severe neurodegenerative disease in brothers with homozygous mutation in POLR1A. Eur. J. Hum. Genet. 2017;25:315–323. doi: 10.1038/ejhg.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jay A.M., Conway R.L., Thiffault I., Saunders C., Farrow E., Adams J., Toriello H.V. Neonatal progeroid syndrome associated with biallelic truncating variants in POLR3A. Am. J. Med. Genet. A. 2016;170:3343–3346. doi: 10.1002/ajmg.a.37960. [DOI] [PubMed] [Google Scholar]
- 56.Rydning S.L., Koht J., Sheng Y., Sowa P., Hjorthaug H.S., Wedding I.M., Erichsen A.K., Hovden I.A., Backe P.H., Tallaksen C.M.E. Biallelic POLR3A variants confirmed as a frequent cause of hereditary ataxia and spastic paraparesis. Brain. 2019;142:e12. doi: 10.1093/brain/awz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minnerop M, Kurzwelly D, Wagner H, Schüle R, Ramirez A. Reply: Biallelic POLR3A variants confirmed as a frequent cause of hereditary ataxia and spastic paraparesis. Brain 142, e13. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.