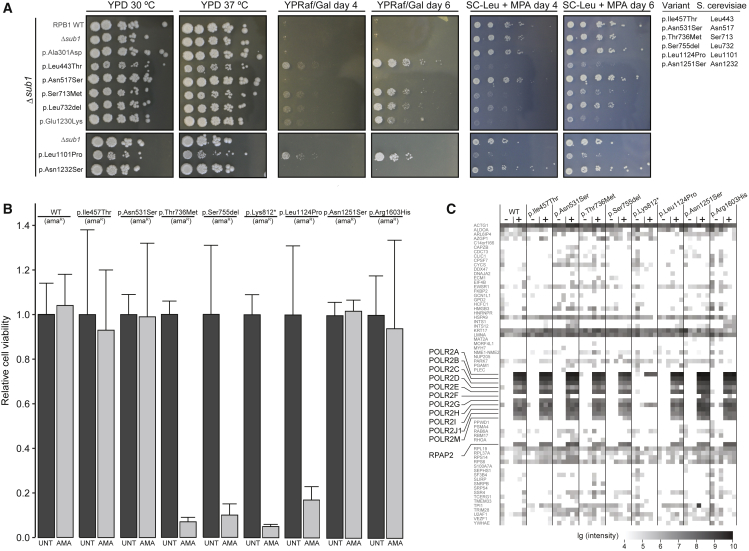
Figure 3.
Functional Evaluation of POLR2A Variants
(A) A growth assay in S. cerevisiae of six rpb1 mutants: p.Ile457Thr (p.Leu443Thr), p.Asn531Ser (p.Asn517Ser), p.Thr736Met (p.Ser713Met), p.Ser755del (p.Leu732del), p.Leu1124Pro (p.Leu1101Pro), and p.Asn1251Ser (p.Asn1232Ser) are compared to wild type (WT). p.Ala301Asp demonstrates a positive control for reduced transcriptional activity, and p.Glu1230Lys demonstrates a positive control for reduced genetic interaction with TFIIS. YPD 30°C depicts growth under normal circumstances, and YPD 37°C depicts growth under stress induced by increased temperature. YPRaf/Gal day 4 and day 6 depict growth under stress induced by increased inappropriate GAL 3′ termination on day 4 and day 6 of culture, respectively. SC-LEU + MPA day 4 and day 6 depict growth under stress induced by the lack of leucine and the addition of mycophenolic acid (MPA) on day 4 and day 6 of culture, respectively.
(B) HeLa cell viability. The individuals’ variants were introduced in an α-amanitin-resistant version of POLR2A (amaR) and expressed from a doxycycline-inducible promoter as a GFP fusion construct in HeLa cells. After the induction of expression, endogenous pol II was left untreated (dark gray bars) or inhibited by the application of α-amanitin (light gray bars), and cell viability was determined. Error bars represent the standard deviation. p.Lys812∗ was used as a representative of p.Gln700∗ and p.Gln735∗ because it is similarly truncated (see Material and Methods).
(C) Interactome analysis of wild-type (WT) and mutant versions of RPB1 using mass spectrometry. Nuclear extracts from HeLa cells expressing GFP-tagged WT or mutant versions of RPB1 were used for precipitations with control agarose beads (−) or GFP-affinity beads (+) in triplicate. In total, 1,133 proteins were identified by more than two peptides via intensity-based absolute quantification (IBAQ) mass spectroscopy. Known members of pol II (highlighted) and a random selection of 5% of the other proteins are shown. The summed peptide intensities of identified proteins were logarithmically transformed to the basis of ten and are represented by shades of gray. Note, intensities smaller than 104 are not obtained due to technical constrains. p.Lys812∗ was used as a representative of p.Gln700∗ and p.Gln735∗.