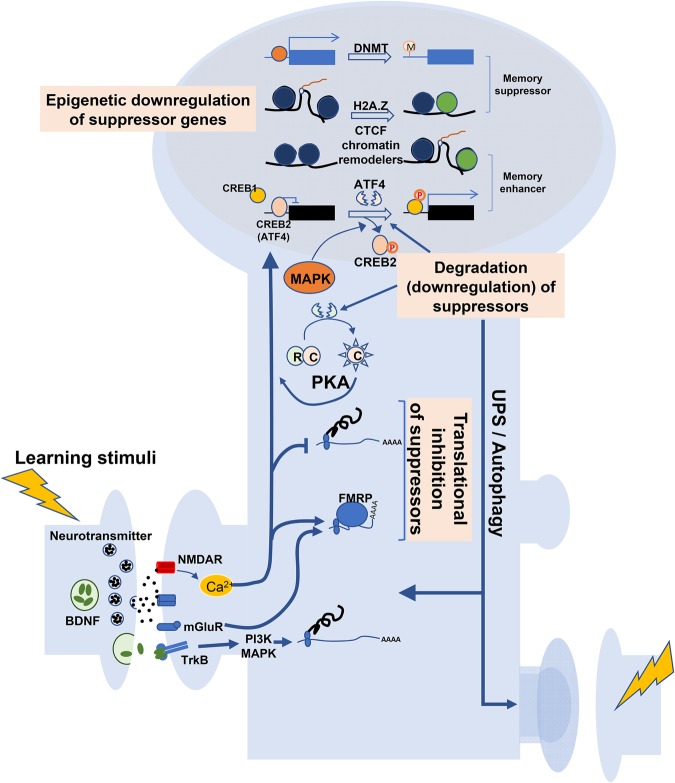
Figure 1.
Schematic diagram showing protein synthesis and degradation involved in memory consolidation. Learning stimuli trigger the presynaptic release of neurotransmitters and BDNF, leading to activation of NMDARs and TrkB receptors, respectively. Secreted BDNF activates TrkB signaling linked with PI3K and MAPK, resulting in increased de novo protein synthesis. Presynaptic neurotransmitters activate NMDARs and subsequent NMDAR-dependent Ca2+ elevation, which not only activates signaling pathways increasing gene transcription and translation but also induces activity-dependent translational suppression of mRNAs that encode memory suppressor proteins, by reducing translation rates or FMRP-dependent inhibition of mRNA translation (Translational inhibition of suppressors). On the other hand, learning stimuli enhance expression and activities of proteolysis mechanisms mediated by UPS or autophagy, leading to degradation of the PKA regulatory subunit (R), ATF4, or other memory suppressor proteins. Degradation of PKA R subunit produces the constitutively active form of PKA catalytic (C) subunit, which triggers diverse molecular mechanisms required for long-term synaptic plasticity. Down-regulation of CREB2 activity is known to induce CREB1-dependent transcription of memory enhancer genes (Degradation or down-regulation of suppressors). Finally, learning stimuli lead to epigenetic down-regulation of memory suppressor genes by suppressing transcription via DNMT-dependent DNA methylation, H2A.Z incorporation to histone complexes, or CTCF-mediated alteration of gene expression. In some cases, such epigenetic modulation was reported to produce an increase in expression of memory enhancer genes, suggesting that epigenetic modulation of transcription is gene-specific (Epigenetic down-regulation of suppressor genes).