Abstract
In spite of the immense progress in hepatitis C virus (HCV) research, efforts to prevent infection, such as generating a vaccine, have not yet been successful. The high price tag associated with current treatment options for chronic infection and the spike in new infections concurrent with growing opioid abuse are strong motivators for developing effective immunization and understanding neutralizing antibodies’ role in preventing infection. Humanized mice—both human liver chimeras as well as genetically humanized models—are important platforms for testing both possible vaccine candidates as well as antibody-based therapies. This chapter details the variety of ways humanized mouse technology can be employed in pursuit of learning how HCV infection can be prevented.
Keywords: Hepatitis C virus, Humanized mice, Immunization, Neutralizing antibodies
1. Introduction
1.1. A Brief History of Hepatitis C Virus (HCV)
Just over three decades ago, hepatitis C virus (HCV) was still an unnamed entity, presumably responsible for the growing number of viral hepatitis cases not caused by the two hepatitis viruses known at the time, hepatitis A (HAV) and hepatitis B (HBV). Even with screening the blood supply for HAV and HBV, cases of transfusion-associated hepatitis not caused by these two viruses were still observed in the mid-1970s [1, 2]. The cause of this disease, nebulously described as non-A-non-B hepatitis (NANBH), remained unknown until the early 1980s when experiments in chimpanzees at least determined that a transmissible agent was in fact at play [3–6]. However, testing for this agent in patients, let alone isolating it, remained impossible until 1989 when ongoing efforts finally resulted in the isolation and characterization of the NANBH virus, now termed HCV [7].
Once identified, assays were rapidly developed to test patients and the blood supply for HCV [8], making blood transfusions safer and allowing for improved epidemiological data regarding the virus’ prevalence. By 1992, the incidence of HCV in the US blood supply was brought down to effectively zero. Since then, treatment options for HCV have undergone numerous evolutions, from the early days of interferon-α2b to pegylated interferon, the advent of ribavirin, and, since 2011, an array of direct-acting antivirals (DAAs). The arsenal of treatments now available for HCV patients have a lower incidence of side effects and cure rates of 90%+ for most genotypes.
1.2. The Continued Need for an HCV Vaccine
While highly encouraging, these achievements in understanding and treating HCV can distract from the fact that HCV continues to remain a potent global health problem, even in the developed nations where so many of these milestone findings occurred. An estimated 71.1 million people worldwide are infected with HCV and are thus at heightened risk for severe liver disease, including cirrhosis, fibrosis, and hepatocellular carcinoma (HCC) [9]. While approximately 15–45% of individuals clear their initial HCV infection, the remaining 55–85% become chronically infected [10–12]. Since the resultant liver disease from chronic infection progresses over many years, patients can go decades after the initial infection event before experiencing symptoms. Thus, these individuals can unknowingly be transmitters during this prolonged period. The Centers for Disease Control and Prevention (CDC) estimates that in the USA alone, 3.5 million people are infected with HCV and anywhere from 40% to 85% of these individuals are unaware [13]. These numbers may even be underestimated due to bias in the populations included in the surveys [14]. Due to the presence of HCV in the blood supply prior to 1992 and the lack of knowledge concerning the virus in the 1970s and 1980s, the CDC has recommended that all individuals born between 1945 and 1965 be tested for HCV as the prevalence of HCV antibodies in these individuals is four times higher than the general population [13].
Besides these increased incidences of chronic HCV infection in the aging baby boomer population, there has been a rise in acute HCV infections amongst a much younger population. Data collected from 2006 to 2012 found a growing number of injection drug users (IDUs)—predominantly white and 30 years or younger living in nonurban areas of central Appalachia—presenting with acute HCV infection upon admission to substance abuse treatment facilities [15]. Amongst IDUs around the world, including in other developed nations, HCV is considered hyperendemic, with mid-point prevalence estimates of HCV antibodies present in 60–80% of IDUs in 25 different countries, including Japan (65%), China (67%), the UK (51%), and the USA (73%) [16]. In fact, mathematical estimates of the number of transmissions stemming from an individual infected with genotypes 1a, 3a, or 4a (i.e., those seen more among IDUs and typical of trends in HCV infection in the Western world) range from 12 to 24.5 [17].
The well-lauded cure now available for HCV is unfortunately not available for all with HCV, underscoring the need for preventive measures such as a vaccine [18]. The extreme cost of these drug regimens—upward of $80,000—makes a worldwide rollout to all HCV patients unfeasible. In the USA, a large percentage of individuals with chronic HCV, such as IDUs and aging individuals infected pre-1992 via blood transfusion, rely on government-funded programs like Medicare and Medicaid. Unable to handle widespread coverage of the exorbitant drug costs, these services have set restrictions, such as sobriety requirements and the need for advanced liver disease progression, to limit patient eligibility [19]. These barriers only add to future healthcare costs as untreated individuals can transmit HCV over a longer period. Furthermore, a focus on curing patients only with severe liver disease is poorly misplaced, as treatment cannot undo the extreme liver damage and probability of developing liver cancer associated with these later stages.
This widespread prevalence of HCV, compounded by the inaccessibility of the most effective treatments, has made a vaccine as necessary as ever before. The holy grail of the HCV field, vaccine development, has faced many challenges, especially since besides humans, HCV only robustly infects chimpanzees. With the increased restrictions on chimpanzee use in biomedical research and the official inclusion of captive chimpanzees on the US Fish and Wildlife Services Endangered Species list, researchers now face additional funding challenges and ethical issues. Thus, to continue HCV vaccine development and related research on neutralizing antibodies (NAbs), immunocompetent small animal models are greatly needed.
1.3. Surrogate Viruses for HCV
Although this chapter will focus on altering mice to support HCV infection and provide a platform for developing vaccines and therapeutics, the inverse approach, whereby HCV or related hepaciviruses are altered to use in small animal models, is also worth noting. Early efforts to find an HCV surrogate utilized the hepacivirus George Barker virus B (GBV-B), which can infect small nonhuman primates such as tamarins [20–23], marmosets [24], and owl monkeys [25]. However, in these animals, GBV-B does not always replicate to robust levels and often causes acute infection instead of the more common persistent infection associated with HCV [24–26]. Chimeric GBV-B viruses containing, for example, parts of the HCV genome encoding the structural [27] or non-structural [28] proteins have had greater success in achieving persistent infection in marmosets, but the utility of such models for vaccine development and drug testing remains unclear.
Over the past few years, enhanced sequencing efforts have identified viruses like HCV that also belong to the Hepacivirus genus of the Flaviviridae family. These related hepaciviruses are naturally occurring in a diverse range of species [29], including horses [30], bats [31], cattle [32, 33], black-and-white colobus monkeys [34], sharks [35], and a variety of rodents [36–38] (but none as of yet in Mus musculus). The genome organization of HCV and these other recently discovered hepaciviruses is similar, but the sequence divergence is substantial. For example, at the amino acid level, rodent hepaciviruses (RHVs) have 70.4% sequence diver-gence from HCV nonstructural proteins, with even greater diver-gence when comparing structural proteins [37]. Very recently, a RHV isolated from wild Norway rats in New York City [38], RHV-nr-1, was successfully used to infect both immunocompromised and immunocompetent laboratory mice, resulting in persistent or acute hepatotropic infection, respectively [39]. Thus, conventional laboratory mice do hold promise for studying the immunological response to hepaciviruses, especially if viral mutants are identified that can establish chronic infection in immunocompetent mice. However, their utility for vaccine development may be limited given the substantial antigenic divergence of these viruses from HCV. Chimeric RHVs could be constructed in which (parts of) the structural and/or nonstructural proteins are replaced, but such approaches have proven difficult as observed for HCV/GBV-B chimeras that are substantially impaired in their replicative fitness. Nonprimate hepaciviruses (NPHVs), which infect horses, are most closely related to HCV, even more so than GBV-B or primate hepaciviruses [34, 37]. However, the obvious difficulties of working in such a nontraditional research model have made NPHVs less than ideal as an HCV surrogate. With the continued efforts to sequence the viromes of diverse species, including other nonhuman primates, it is possible that more hepaciviruses will be uncovered. Until then, other approaches have focused on viral adaptation to facilitate study of HCV infection in mice. For example, the HCV strain Jc1 (genotype 2a) was successfully adapted to use murine CD81 via mutations in E1 and E2, permitting viral uptake in murine hepatocytes both in vitro and in vivo [40, 41].
1.4. Humanizing Mice to Study HCV
Mice have a long history of use in biomedical research, but they naturally do not support HCV infection. However, the plethora of tools and reagents available to study these animals and the relative ease in performing genetic manipulations have made mice suitable candidates for a variety of humanization procedures. This has allowed researchers to not only infect humanized mice with HCV but has also created new opportunities for studying vaccines and broadly neutralizing antibodies (bNAbs) in vivo. In this chapter, we will broadly describe the approaches available for generating genetically humanized mice or human liver chimeric mice and more specifically the ways to perform and assess HCV infection in such models. As vaccination regimens and antibody treatments are highly individualized to specific experiments, we will present an overview of how these methods have been successfully utilized to test HCV immunization approaches in these models.
2. Materials
Before proceeding with any work involving humanized mice and/or human pathogens, please see Note 1.
2.1. Mouse Strains
Note that all mice expressing HCV entry factors, specifically human CD81, OCLN, SCARB1 and CLDN1 should be on a reporter background enabling activation of a cellularly encoded luciferase gene that can be activated by Cre recombinase (i.e., FVB.129S6 (B6)-Gt(ROSA)26Sortm1(Luc)Kael/J [42]).
Expression of HCV entry factors in these mice can be accomplished in various ways:
2.2. BiCre-Jc1 Generation
pBi-nlsCre-Jcl construct (described in [44]).
RNeasy mini kit (Qiagen).
T7 RiboMAX™ Express Large-Scale RNA production system (Promega).
Huh-7.5.1 cells (Chisari laboratory, The Scripps Research Institute [47]).
BTX ECM 830 electroporator (Harvard Apparatus).
Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific).
Stirred cell (Millipore).
0.45 μm bottle-top filter (Millipore).
2.3. IVIS
IVIS Lumina II platform (Caliper Lifesciences).
d-Luciferin salt (Gold Biotechnology).
0.2 μm filter (Thermo Fisher Scientific).
Isoflurane (TW Medical).
28G × ½ insulin syringes (BD Biosciences).
2.4. Quantifying HCV RNA
ZR Viral RNA kit (Zymo Research).
Nuclease-Free Water (Luminex).
HCV primers for PCR (ordered from Integrated DNA Technologies).
Reverse Transcriptase (Luminex).
MultiCode ISOlution (Luminex).
Titanium Taq (Clontech).
HCV standard (homemade; RNA in vitro-transcribed from the plasmid, pIDTBlue, containing the Jc1-RNA sequence “GGCUCCAUCUUAGCCCUAGUCACGGCUAGCU-GUGAAAGGUCCGUGAGC” as cloned by Integrated DNA Technologies).
LightCycler 480 Real-Time PCR System (Roche Applied Sciences).
MultiCode Analysis Software v1.6.5 (Luminex).
3. Methods
3.1. Genetically Humanized Mice Can Support HCV Entry
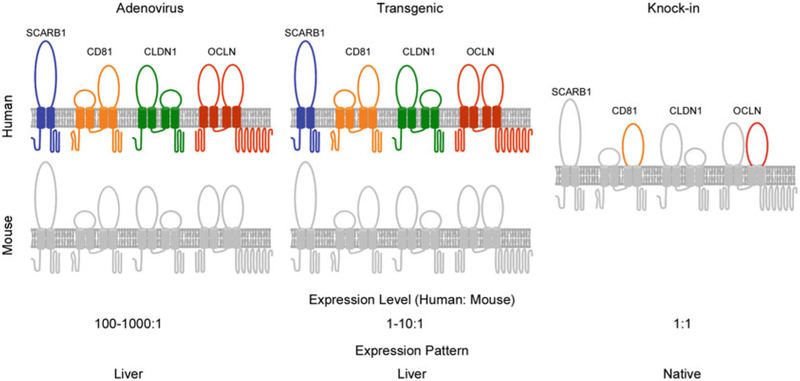
Humanized mice for HCV research are generally created in one of two ways: by expression of human factors known to be necessary for part(s) of the HCV life cycle (Fig. 1) or by xenotransplantation. In murine cells, HCV replicons are able to replicate, but uptake of cell-culture produced virus or HCV pseudoparticles is not possible. Human CD81 [48], scavenger receptor class B type I (SCARB1) [49], and claudin-1 (CLDN1) [50] were all known human entry factors for HCV, but it was the additional identification of human occludin (OCLN) [51] as an essential entry factor. The combined expression of these four factors, or at a minimum human CD81 and human OCLN, finally led to observed uptake of HCV in murine cells in vitro [51]. This work was expanded in vivo to genetically humanize mice by adenoviral delivery of human OCLN and human CD81 [43, 44], allowing for HCV uptake in the liver of a fully immunocompetent animal. However, viral replication and infectious particle production were not observed in either these mice or those subsequently engineered to transgenically express human CD81 and human OCLN in the murine liver under the control of an albumin promoter [52]. Only upon blunting the innate immune response of these entry factor transgenic mice was the complete HCV life cycle observed as evidenced by viral RNA in both the serum and liver [52].
Fig. 1.
Summary of genetically humanized mice supporting HCV uptake. The minimal factors required for HCV uptake in human hepatocytes are CD81, SCARB1, CLDN1, and OCLN, with human CD81 and OCLN constituting the minimal set of human factors for entry into murine hepatocytes. Thus, fully immunocompetent mice can support viral uptake by expressing these factors by adenoviral delivery or by genetic manipulation. The latter approach has been further utilized in two ways, transgenic expression of hCD81 and hOCLN under the controlof an albumin promoter or knockin of humanized CD81 and OCLN where the second extracellular loops of the murine orthologs of these two proteins have been altered to facilitate HCV entry. While all these methods have proven successful, the knockin model best matches the natural tissue expression and physiological levels of CD81 and OCLN, which are expressed only in the liver and at supraphysiological levels in adenovirally transduced or transgenic mice
To express HCV entry factors by adenovirus, high particle numbers are delivered to mice (in this case, a Cre reporter strain of choice as described below) with the goal of widespread expression of these factors in the murine hepatocytes. cDNA encoding human CD81, SCARB1, CLDN1 or OCLN can been cloned into individual pAdEasy AV5 adenoviral vectors (Agilent) (for details see [43]). Alternatively, instead of expressing each factor in individual vectors, the cDNA of CD81 and SCARB1 or CLDN1 and OCLN can instead be linked by a sequence encoding the 2A self-cleavage peptide to ensure the four factors are separate proteins following translation [53].
Although these models have been extremely useful for studying HCV infection in vivo, CD81 and OCLN are expressed only in hepatocytes and at supraphysiological levels. To overcome these drawbacks, knockin mice were recently generated that harbor CD81 and OCLN alleles minimally humanized so that the second extracellular loops of these two proteins match the relevant human sequence important for HCV entry [46]. These mice develop normally without apparent defects, express CD81 and OCLN at physiological levels across a variety of tissues, and support uptake of HCV to a similar extent as in the two models described above.
In stark contrast to the above studies in genetically humanized mice, where completion of the HCV life cycle required a dampened innate immune response, one group has successfully demonstrated persistent HCV infection in genetically humanized mice with an intact immune response. Here, human CD81 and OCLN were once more expressed under an albumin promoter but in an ICR background [45]. As genetically humanized immunocompetent C57BL/6 mice have not previously become viremic in similar experiments [44, 52], the authors point to differences in the expression of interferon-stimulated genes (ISGs) and host factors such as ApoE and miR-122 upon HCV infection as possibly responsible. While these findings are striking, genetic manipulations of ICR mice, an outbred strain, present additional technical challenges that further research will need to overcome.
3.2. Human Liver Chimeric Mice for Studying the Complete HCV Life Cycle
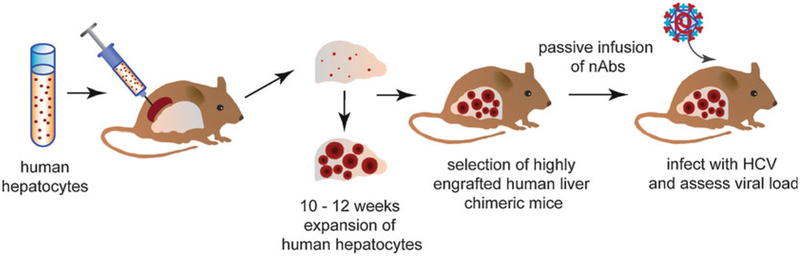
The second broad category of mouse humanization relevant to HCV research involves xenotransplantation to generate human liver chimeric mice (Fig. 2) with or without human immune system reconstitution. In order to accept engrafted tissue, these mice must be immunocompromised and must also have a hepatic environment conducive to the proliferation of engrafted cells over endogenous murine cells. The latter condition can be met by treating the mice with chemical agents such as tetrachloride or retrorsine, or, more typically, by changes at the genetic level. Genetic manipulation was first successfully used to induce liver failure in a mouse strain by murine urokinase plasminogen activator overexpression driven by the albumin promoter (Alb-uPA) [54]. When backcrossed with a severe combined immunodeficiency strain (SCID), the resultant SCID/Alb-uPA mice could be stably engrafted with human hepatocytes that supported HCV infection [55]. To overcome some of the disadvantages of this model, including the limited period when engraftment can be performed, a strain where the major urinary protein (MUP) promoter drives uPA expression is backcrossed with a SCID strain to form SCID/MUP-uPA mice that can undergo successful engraftment and HCV infection [56].
Fig. 2.
The generation of human liver chimeric mice for passive immunization against HCV
Knockout of the liver enzyme fumaryl acetoacetate hydrolase (FAH) has also been pursued as a method for causing selective death of endogenous murine hepatocytes. To permit actual engraftment, FAH−/− mice were crossed with Rag2−/− IL-2RγNULL mice to create FNRG mice [57]. The absence of FAH specifically in the murine hepatocytes results in a buildup of toxic metabolites during tyrosine catabolism leaving the engrafted human hepatocytes, which still express FAH, at a growth advantage. Importantly, the FAH deficiency in murine hepatocytes can be overcome by treatment with the compound 2(2-nitro-4-trifluoromethylbenzoyl)-1,3 cyclohexanedione (NTBC). NTBC cycling thus permits “tunable” liver injury throughout the mouse’s lifespan (for detailed information about generating and maintaining these mice see [58]). The level of engraftment corresponds to human albumin levels in the mouse’s serum as determined by ELISA. Similarly, immunodeficient NOG mice expressing the transgene herpes simplex virus type 1 thymidine kinase (HSV-tk) can be treated with ganciclovir for targeted destruction of murine hepatocytes, thus allowing engrafted human hepatocytes to proliferate instead [59]. This model has also been successfully infected with HCV [60].
These numerous models permit both the stable engraftment of adult human hepatocytes in mice as well as sustained HCV infection. However, the immunocompromised nature of these mice has limited their utility for studying immune responses to HCV infection and their subsequent impact on pathogenesis. Toward this aim, dually reconstituted mice have also been generated with engraftment of both human hepatocytes and components of a human immune system [61–67]. However, widespread use of these dually engrafted mice has been limited due to variation across donors, difficulty in acquiring syngeneic human HSCs and hepatocytes, the required technical expertise, and high cost.
3.3. Assessing HCV Entry in Genetically Humanized Mice
Although HCV does not replicate in genetically humanized mice unless the innate immune response is blunted, viral entry is still supported. The ability to detect uptake of HCV in these mice is necessary to assess the efficacy of NAbs or vaccines on this step of the viral life cycle. Mouse lines containing a loxP-flanked stop cassette upstream of a marker (i.e., firefly luciferase) are readily available from commercial vendors such as the The Jackson Laboratory (Bar Harbor, see below). In the presence of Cre recombinase, the transcriptional stop cassette undergoes recombination and is effectively removed, allowing marker expression to now occur. Recombinant HCV expressing Cre recombinase can be generated (see Subheading 3.3.1) and injected into humanized Cre reporter mice. While below we describe the use of a genotype 2a chimera expressing Cre recombinase, this model can also be used to test HCV Cre expressing the envelope proteins of other genotypes. Upon entry of the virus, the translation of the HCV polyprotein will release the Cre recombinase, which can now move to the nucleus and facilitate recombination of the transcriptional stop cassette. In this way, hepatocytes that uptake virus will now express the marker of interest, which, in cases such as firefly luciferase, can be detected in vivo.
Examples of commonly used reporter strains available from The Jackson Laboratory (Bar Harbor ME) are FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael/J (also known as R26-LSL-FLuc) [42] and B6;129-Gt(ROSA)26Sortm1Joe/J (also known as R26-LSL-GNZ) [68] mice which, following activation by Cre recombinase, drive expression of firefly luciferase or a nuclear EGFP and β-galactosidase fusion protein, respectively.
3.3.1. Production of Bicistronic HCV Expressing Cre Recombinase
The sequence of the genotype 2a HCV chimera J6/JFH1 (also known as Jc1) is deposited (GenBank accession number: JF343782) and the pBi-nlsCre-Jc1 construct is available upon request upon completion of appropriate material transfer agreements.
Digest 10 μg of the pBi-nlsCre-Jc1 construct with XbaI to linearize the construct for in vitro transcription to produce pBi-nlsCre-Jc1 RNA. To ensure complete linearization, assess 5% volume of the digested plasmid by agarose gel electrophoresis.
Purify the linearized BiCre-Jc1 plasmid and use 1 μg to pro-duce pBi-nlsCre-Jc1 RNA by in vitro transcription (T7 RiboMAX Express Large Scale RNA Production System, Promega).
Purify the pBi-nlsCre-Jc1 RNA using method of choice and quantify the concentration of viral RNA by NanoDrop.
As a quality control test, let RNA sit for 30 min on wet ice before running a few microliter on an agarose gel to check for degradation (see Note 2). If RNA is of sufficient integrity, prepare 5 μg aliquots and store at −80°C for future use, only thawing on wet ice immediately before electroporation.
As all subsequent steps involve virus production, necessary biosafety precautions must be taken (see Note 3). Electroporate the pBi-nlsCre-Jc1 RNA into Huh-7.5.1 cells (DMEM containing 5% FBS) as previously described in [43]. As an additional control for electroporation efficiency, seed in a 6-well plate some cells electroporated with or without viral RNA. Collect these cells 72 h post-electroporation and assess the percentage of cells positive for NS5A by flow cytometry.
On the plates of electroporated cells, change the medium to DMEM without FBS 24 h post-electroporation. Collect supernatants every 6 h starting from 72 h until 144 h, always replacing the media with serum-free DMEM. Supernatants can be pooled and stored at 4°C in a foil-covered container.
Filter the pooled supernatants through a 0.45 μm bottle top filter (Millipore). Store at least 1 mL of this unconcentrated virus at −80°C to have as a control for concentration efficiency in step 8. Concentrate the remaining filtered supernatant using a stirred cell (Millipore). Aliquot the concentrated virus and store at −80°C for further use.
Thaw an aliquot of both concentrated and unconcentrated virus to determine the viral titer (TCID50) on Huh7.5 cells as previously described in [69].
3.3.2. Bioluminescence Imaging to Assess HCV Entry In Vivo
In vivo bioluminescence imaging allows for rapid, minimally invasive, longitudinal quantification of reporter gene activation in mice expressing at least the minimal HCV entry factors and subsequently infected with Bi-nlsCre-Jc1. Luminescence measurements generally ensure greater sensitivity through deep tissues and the skin. Here, we outline bioluminescence measurements specifically using the IVIS Lumina II platform (Caliper Life Sciences) of mice following adenoviral delivery of the four canonical human entry factors and subsequent infection with HCV Cre (equivalent imaging plat-forms from other manufacturers could conceivably be used). The IVIS Spectrum imager expresses the bioluminescent signal in photons per second and displays it as an intensity map. The luminescence, which is the consequence of the photon flux emitted by the luciferase-expressing cells, can be measured using a region of interest (ROI) tool.
Prepare stock solution of d-Luciferin at 15 mg/mL in DPBS (no calcium, no magnesium). Filter-sterilize through a 0.2 μm filter, aliquot, and store at −20°C for future use.
Inject intravenously Rosa26-LSL-Fluc mice with an equal mixture of adenovirus encoding all four HCV human entry factors (1011 adenoviral particles per entry factor).
24 h following adenoviral entry factor delivery, inject mice intravenously with 2 × 107 TCID50 Bi-nlsCre-Jc1.
72 h following injection of Bi-nlsCre-Jc1, prepare the animal for in vivo bioluminescence imaging. Anaesthetize the animal using an isoflurane vaporizer. Inject intraperitoneally appropriate volume (typically ~200 μL) of sterile 15 mg/mL d-luciferin stock solution (see step 1) so that each animal receives 150 mg d-Luciferin/kg body weight (equivalent to 10 μL of d-luciferin stock solution per gram of body weight). Place mouse in dorsal recumbency (abdomen facing up) inside the camera box of the IVIS Spectrum imager.
Measure the bioluminescence signal using the IVIS Lumina II in vivo bioimaging system. Due to in vivo uptake of d-Luciferin over time, capture sequential images of the mice every 5 min until luminescence saturation is reached.
3.4. Quantifying HCV RNA in Human Liver Chimeric Mouse Serum
In the case of human liver chimeric mice, reporter genomes are not necessary since HCV can robustly replicate to levels detectable in both the liver and serum of a well-engrafted animal. Thus full-length genomes such as JFH1 (genotype 2a) can be used for infections. HCV replication over time is easily tracked from relatively small blood volumes obtained by submandibular bleedings. Additionally, viral load can be assessed in the mouse’s liver but requires perfusion of the organ as described in [58] and subsequent RNA isolation from a kit compatible for use with animal tissues. Once the RNA has been isolated, the same procedure detailed below for RT-qPCR of HCV RNA isolated from animal serum can be used. The method given below is one approach for viral RNA quantification specifically using EraGen MultiCode-RTx Technology (Era-Gen Biosciences).
Draw approximately 200 μL of whole blood by submandibular bleeding into a 1.5 mL microcentrifuge tube.
Allow the blood to sit upright untouched for 10–20 min at room temperature for clotting.
Centrifuge the blood for 10 min, 3000 rpm (956 × g), 4°C. Following centrifugation, there should clearly be two layers in the tube. Transfer the upper layer (serum) to a fresh 1.5 mL microcentrifuge tube (see Note 4).
Purify RNA from the plasma using the ZR Viral RNA kit (Zymo Research) or an equivalent kit.
Quantify the viral RNA copy number by one step RT-qPCR using EraGen MultiCode-RTx Technology (EraGen Biosciences). The following primers were used for the detection of HCV RNA: 5′-GCTCACGGACCTTTCA-3′ (sense) and 5′-GGCTCCATCTTAGCCC-3′ (antisense). For use with EraGen MultiCode-RTx Technology, the sense primer requires the following modifications, which can be specified upon ordering from oligonucleotide providers such as Integrated DNA Technologies at the 5′ end: 6-FAM and int 5-Methyl dC (iMe-dC).
Prepare the reaction for each sample as in Table 1, making sure to also prepare serial dilutions of an HCV standard to generate a standard curve.
Run the reactions on a LightCycler 480 Real-Time PCR System (or equivalent device) using the thermal profile in Table 2.
Import the multicomponent data from the run into the MultiCode Analysis Software v1.6.5 for analysis.
Table 1.
Reaction conditions for HCV RNA RT-qPCR
| Reagents | Volume (μL) |
|---|---|
| Nuclease-free H2O | Up to 25 |
| 5 × MultiCode ISOlution | 5 |
| MMLV reverse transcriptase | 0.5 |
| Primer mix (5 μM forward + 5 μM reverse in nuclease-free-H2O) | 1 |
| RNA | x |
| Titanium Taq | 0.5 |
Table 2.
Cycling conditions for quantifying HCV RNA in serum
| Procedure | Temperature, °C | Time |
|---|---|---|
| Reverse transcription | 50 | 5 min |
| Hot start | 95 | 3 min |
| 50 cycles | 95 | 5 s |
| 58 | 10 sa | |
| 72 | 20 s | |
| Melt Curve | 95 | 15 s |
| 60 | 1 min | |
| 95 | 15 sa |
Data collection must be turned on at these steps for proper analysis in step 8
3.5. Humanized Mice for Designing and Testing HCV Vaccinesand Therapeutic Antibodies
As genetically humanized mice are still immunocompetent and do not support the full HCV life cycle, vaccine and therapeutic anti-body studies utilizing these models have largely focused on whether viral uptake can be reduced. In contrast, immunodeficient human liver chimeric mice have primarily been used for passive immunization studies in which animals are infused with antibodies directed against viral antigen(s). Both prophylactic and therapeutic studies have been performed in human liver chimeric mice challenged with HCVcc or patient-derived sera. The antibodies used for passive immunization, as described in several examples below (summarized in Table 3), have been isolated from patient sera or from immunocompetent mice injected with HCV or viral components like the envelope proteins E1 and/or E2.
Table 3.
Summary of HCV immunization studies performed in humanized mice
| Expression approach | Virus | Immunization | |
|---|---|---|---|
| Genetically humanized mice | Adenovirus | HCV-Cre expressing genotypes 1b, 2a, or 4a structural proteins [43] | Active |
| In vitro neutralization assay with chimeric HCVcc panel covering all seven HCV genotypes; BiCre-Jc1 (genotype 2a) in vivo [52] | Active | ||
| HCV-Cre recombinase expressing structural proteins of Con1 (genotype 1b) or Jc1 (genotype 2a) [80] | Passive | ||
| BiCre-Jc1 (genotype 2a) [102] | Passive | ||
| Mouse | Virus | Immunization | |
| Human liver chimeric mice | uPA/SCID | Heterologous HCV inoculum from an HCV genotype 1a infected patient [94] | Passive |
| Patient-derived genotype 1b [96] | Passive | ||
| H77C (genotype 1a), ED43 (genotype 4) [104]) | Passive | ||
| Prevention experiments: genotype 1b or genotype 4 HCV-infected serum; treatment experiments: Jc1 (genotype 2a), VL-JFH1 (genotype 1b/2a chimera), genotype 2a serum, genotype 4 serum [106] | Passive | ||
| J6/JFH1 (genotype 2a) [107] | Passive | ||
| Mouse-passaged H77C (genotype 1a) [108] | Passive | ||
| FNRG PXB-mouse (cDNA-uPA/SCID) | H77 (genotype 1a), J6/JFH1 (genotype 2a) [102] | Passive | |
| H77S (genotype 1a) [103] | Passive | ||
| Mouse-passaged HCR6 (genotype 1b) [105] | Passive | ||
3.5.1. Active Vaccination
As described above, genetically humanized mice remain immunocompetent and can thus be utilized for active immunization studies. One such approach for active immunization utilizes recombinant vaccinia virus (rVV) as a vector to deliver viral genes. Previous work showed that large pieces of pathogen-derived DNA could be easily inserted into the VV genome without disrupting viral replication [70, 71]. rVVs expressing different HCV genes have been tested in chimpanzees [72–74] and mice [75, 76]. In genetically humanized mice expressing human CD81 and human OCLN, injection with a rVV expressing HCV genotype 1a C-E1-E2-p7-NS2 resulted in robust antibody production against HCV E2 and reduced viral entry following subsequent challenge with different HCV genotypes [44]. An alternative to injecting a viral vector to express viral proteins is to inject the viral protein itself. Previous efforts to cost-effectively produce large-scale amounts of viral protein for clinical applications and the poor results of recombinant vaccine trials in humans had made this less appealing. However, a soluble form of E2 with a truncated transmembrane domain (sE2) was recently generated that induced NAbs in immunocompetent mice with broad, pan-genotypic activity in vitro and could be produced to scale in insect cells [53]. Furthermore, genetically humanized mice were protected against viral challenge following sE2 immunization.
Active immunization studies in mice, as in other species, can determine the type and magnitude of antibody response generated as well as the potential protection provided against subsequent viral challenge. In vitro neutralization assays can be performed using the sera of mice, which do not have to be genetically humanized, collected multiple weeks after immunization to insure sufficient time for antibody production. Sera can then be incubated with JFH1 chimeras expressing the structural proteins from multiple HCV genotypes to test for their range of neutralization. Additionally, immunized mice can be subsequently challenged with HCV in vivo. Here immunization must be performed in one of the reporter mouse strains described above and an appropriate HCV genome used (i.e., BiCre-Jc1) in order to have a readout of viral uptake since detectable replication levels in the liver and/or serum are negligible in these immunocompetent models.
3.5.2. Neutralizing Antibodies
Many of the efforts to identify NAbs capable of neutralizing different HCV genotypes predate the use of humanized mouse models. In patients, there has been widespread debate about how NAbs may affect HCV replication and the nature of their contribution to viral clearance in a given individual (reviewed in [77]). Thus, better understanding NAbs has been of great interest in designing both prophylactic and therapeutic HCV treatments. With as much as 35% difference between the sequences of HCV genotypes [78], the so-called broadly NAbs, able to act against multiple genotypes or even the variety of quasispecies present in a single infected individual, are of particular interest. This sequence diversity also under-scores the need for HCV treatments with broad, pangenotypic activity to lower the risk of viral escape mutants. Prior reliance on in vitro systems has only added to the controversy concerning NAbs in patients. Thus, the advent of humanized mouse research has provided an additional platform for exploration of NAbs in the context ofHCV infection.
Repeated work has shown that HCV NAbs primarily target the mediators of viral entry, the HCV envelope proteins E1 and E2 [79–87]. Thus, many efforts to induce a strong NAb response have focused on using recombinant HCV protein(s), primarily E1 and/or E2, to generate vaccines. Prior to the availability of humanized mice in the HCV field, such vaccines were tested in both nonhumanized rodent models [88–90] and humans [91]. The failure of the human recombinant vaccine trials to induce a robust NAb response [91–94] and the rise in humanized mouse models for HCV research has heightened interest in both generating better recombinant vaccines and understanding NAbs.
Many promising NAbs previously identified in other platforms have been re-examined in human liver chimeric mice. One such study found candidates able to bind HCV E2 following an extensive search of a phage-display library of antibody antigen-binding fragments (Fabs) derived from a chronically infected HCV patient. Select Fabs were used to generate recombinant monoclonal anti-bodies (MAbs) that were further tested in vitro. The most promising candidates from these assays specifically bound antigenic region 3 (AR3A and AR3B) of the HCV envelope glycoprotein E2, and injection of these candidates into human liver chimeric (Alb-uPA/SCID) mice was able to protect some from infection with a heterologous HCV inoculum from an HCV genotype 1a infected patient [95]. Prophylactic administration of high concentrations (200 mg/kg) of antibody AR3A or AR4A was also found to protect genetically humanized mice against challenge with genotype 1b or genotype 2a cell-culture produced HCV (HCVcc) [81]. AP33 [87, 96] and 3/11 [80], which were identified prior to the advent of humanized mice in rodents immunized with E1 and/or E2, were also tested for their ability to protect against challenge with patient-derived HCV (genotype 1b) in Alb-uPA/SCID mice [97]. Although AP33 and 3/11 recognize overlapping epitopes in E2, only AP33 (at 100 mg/kg) demonstrated the ability to protect the humanized mice from infection. In line with this work and the growing use of humanized mice for such studies, efforts have also been made to better characterize the antigenic regions of full-length and soluble E1E2 and the antibody responses they elicit specifically in mice [98]. Furthermore, vectored immunoprophylaxis, whereby adenoviruses are used to deliver NAbs, has been an approach for mouse studies of pathogens such as HIV, Plasmodium falciparum, and influenza [99–102]. This method has also been performed in genetically humanized mice expressing the human HCV entry factors as well as in liver chimeric FNRG mice [103]. Whether supplied individually or in combination, the NAbs tested were able to block HCV entry in the genetically humanized mice and to prevent viremia in liver chimeric FNRG mice following viral challenge. Furthermore, in this same study, liver chimeric mice already infected with J6/JFH (genotype 2a) were therapeutically treated with NAbs, leading to a drop in HCV RNA levels below the limit of detection in the sera.
3.5.3. Host Factors as Antibody Targets
While the majority of work has focused on antibodies directed against viral targets, some studies have tested the potential for antibodies against host factors as a short-term treatment for patients undergoing liver transplantation. Directed against a necessary HCV entry factor, anti-CD81 mAbs (K04 [104] and JS81 [105]) have been tested for both their prophylactic and therapeutic abilities in human liver chimeric mice. With frequent dosing, K04 treatment started post-HCV inoculation could substantially abrogate infection. Prophylactic use of either antibody did effectively protect from infection, but a high dose of Ab was required and effects on mouse health were observed.
Anti-CLDN1 mAbs have also proven successful as prophylactic treatment against HCV infection (mouse-passaged genotype 1b, 104 RNA copies) without any significant side effects in human liver chimeric mice [106]. A similar study demonstrated prophylactic treatment of human liver chimeric mice with a different anti-CLDN1 mAb followed by a “booster” 1 and 5 days post-infection resulted in undetectable HCV RNA in the serum for six weeks following infection [107]. Even as the concentration of antibody diminished in the sera of these mice, infection with either genotype 1b or genotype 4 sera were prevented. Therapeutic administration of anti-CLDN1 mAb was also successful in cohorts of human liver chimeric mice chronically infected with one of a variety of HCV genotypes (HCVcc Jc1; genotype 2a serum; neutralization escape variant HCVcc VL-JFH1; genotype 4). Importantly, this CLDN1-specific mAb displayed no major toxicity or adverse effects in the mice.
3.5.4. IgG as a Prophylactic
UV-inactivated HCVcc (J6/JFH1) has also been used immunize immunocompetent BALB/c mice (four separate injections at two week intervals), resulting in production of anti-E1 and anti-E2 antibodies [108]. Purified IgG from the sera of these mice was subsequently used as a prophylactic vaccine in Alb-uPA/SCID mice 1 week before infection with HCVcc (J6/JFH1). Infection was effectively inhibited at low doses (103 HCV RNA copies), but not at higher challenges of 104 or 105 RNA copies. Broader approaches have also been taken using anti-HCV immunoglobulin G (IgG) obtained from a chronically infected patient in Alb-uPA-SCID mice to assess its ability to protect against challenge with a genotype 1a clone derived from the acute stage of infection in the same patient [109]. Passive immunization with the patient IgG was able to confer protection against viral challenge, albeit with an extremely homogeneous inoculum.
3.6. Conclusions and Future Outlook
In this chapter, we described the numerous applications of humanized mice for assessing potential HCV vaccines and therapeutic antibodies. While this work has continued to push the field forward, several limitations of these models remain. Studies in immunocompetent, genetically humanized mice focus on humoral immunity in the context of an HCV infection that is restricted to viral uptake. Genetic humanization allows for a relatively limited number of human factors to be expressed. The ever-lengthening list of HCV entry factors thus present in an actual patient may provide additional challenges that the mouse model cannot fully recapitulate. For human liver chimeric mice, which cannot amount any kind of adaptive immune response due to their immunocompromised status, conclusions are also limited. However, efforts to combine the benefits of human liver chimeric mice—i.e., studying HCV infection past entry into hepatocytes—with the presence of a humanized immune system are ongoing. This work will aid in filling the gaps of current models and create a more accurate picture of the complex relationship between HCV and the host immune response. Furthermore, improved dually reconstituted mice will greatly strengthen our knowledge of immunopathogenesis and the downstream liver disease observed in chronically infected patients
4. Notes
Researchers must receive approval from their Institutional Animal Care and Use Committee (IACUC), Institutional Review Board (IRB), Institutional Biosafety Committee (IBC), or the equivalent institutions before proceeding with work involving humanized mice and/or human pathogens.
When running the in vitro-transcribed RNA on a gel, it is highly recommended to also run a 1 kb ladder. If the RNA is of good quality, a bright, single band should appear at around 3 kb. Beware of using RNA with excessive smearing as this is likely due to degradation and will result in decreased efficiency of subsequent steps.
All virus work should be performed in a BSL safety level 2 hood. Following electroporation, all cuvettes and tips that came into contact with electroporated cells need to soak in Vesphene for at least 20 min before disposal.
Around 200 μL of whole blood should yield at least 50 μL of plasma, which is a sufficient volume for the protocol described here.
Acknowledgments
This study is supported by grants from the National Institutes of Health (R01 AI079031, R01 AI107301, R21AI117213 to A.P.), a Research Scholar Award from the American Cancer Society (RSG-15-048-01-MPC to A.P.), a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (to A.P.) and funds from Princeton University. J.M.G. was in part supported by cofunding from NIAID on iNRSA 5T32GM00738 and Q.D. by a postdoctoral fellowship from the New Jersey Commission on Cancer Research.
References
- 1.Alter HJ, Holland PV, Morrow AG, Purcell RH, Feinstone SM, Moritsugu Y (1975) Clinical and serological analysis of transfusion-associated hepatitis. Lancet 2:838–841 [DOI] [PubMed] [Google Scholar]
- 2.Feinstone SM, Kapikian AZ, Purcell RH, Alter HJ, Holland PV (1975) Transfusion-associated hepatitis not due to viral hepatitis type A or B. N Engl J Med 292:767–770 [DOI] [PubMed] [Google Scholar]
- 3.Alter HJ, Purcell RH, Holland PV, Popper H (1978) Transmissible agent in non-A, non-B hepatitis. Lancet 1:459–463 [DOI] [PubMed] [Google Scholar]
- 4.Bradley DW, Cook EH, Maynard JE, McCaustland KA, Ebert JW, Dolana GH et al. (1979) Experimental infection of chimpanzees with antihemophilic (factor VIII) materials: recovery of virus-like particles associated with non-A, non-B hepatitis. J Med Virol 3:253–269 [DOI] [PubMed] [Google Scholar]
- 5.Hollinger FB, Gitnick GL, Aach RD, Szmuness W, Mosley JW, Stevens CE et al. (1978) Non-A, non-B hepatitis transmission in chimpanzees: a project of the transfusion-transmit-ted viruses study group. Intervirology 10:60–68 [DOI] [PubMed] [Google Scholar]
- 6.Tabor E, Gerety RJ, Drucker JA, Seeff LB, Hoofnagle JH, Jackson DR et al. (1978) Transmission of non-A, non-B hepatitis from man to chimpanzee. Lancet 1:463–466 [DOI] [PubMed] [Google Scholar]
- 7.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M (1989) Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362 [DOI] [PubMed] [Google Scholar]
- 8.Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH et al. (1989) An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362–364 [DOI] [PubMed] [Google Scholar]
- 9.Polaris Observatory HCV Collaborators (2017) Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2(3):161–176 [DOI] [PubMed] [Google Scholar]
- 10.Thomson EC, Fleming VM, Main J, Klenerman P, Weber J, Eliahoo J et al. (2011) Predicting spontaneous clearance of acute hepatitis C virus in a large cohort of HIV-1-infected men. Gut 60:837–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlach JT, Diepolder HM, Zachoval R, Gruener NH, Jung MC, Ulsenheimer A et al. (2003) Acute hepatitis C: high rate of both spontaneous and treatment-induced viral clearance. Gastroenterology 125:80–88 [DOI] [PubMed] [Google Scholar]
- 12.WHO (2016) Guidelines for the screening care and treatment of persons with chronic hepatitis C infection: updated version WHO guidelines approved by the guidelines review committee. WHO, Geneva: [PubMed] [Google Scholar]
- 13.Smith BD, Morgan RL, Beckett GA, FalckYtter Y, Holtzman D, Teo CG et al. (2012) Recommendations for the identification of chronic hepatitis C virus infection among persons born during 1945–1965. MMWR Recomm Rep 61:1–32 [PubMed] [Google Scholar]
- 14.Edlin BR (2011) Perspective: test and treat this silent killer. Nature 474:S18–S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zibbell JE, Iqbal K, Patel RC, Suryaprasad A, Sanders KJ, Moore-Moravian L et al. (2015) Increases in hepatitis C virus infection related to injection drug use among persons aged </ =30 years-Kentucky, Tennessee, Virginia, and West Virginia, 2006–2012. MMWR Morb Mortal Wkly Rep 64:453–458 [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson PK, Mathers BM, Cowie B, Hagan H, Des Jarlais D, Horyniak D et al. (2011) Global epidemiology of hepatitis B and hepatitis C in people who inject drugs: results of systematic reviews. Lancet 378:571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magiorkinis G, Sypsa V, Magiorkinis E, Paraskevis D, Katsoulidou A, Belshaw R et al. (2013) Integrating phylodynamics and epidemiology to estimate transmission diversity in viral epidemics. PLoS Comput Biol 9: e1002876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Callaway E (2014) Hepatitis C drugs not reaching poor. Nature 508:295–296 [DOI] [PubMed] [Google Scholar]
- 19.Trooskin SB, Reynolds H, Kostman JR (2015) Access to costly new hepatitis C drugs: medicine, money, and advocacy. Clin Infect Dis 61:1825–1830 [DOI] [PubMed] [Google Scholar]
- 20.Beames B, Chavez D, Lanford RE (2001) GB virus B as a model for hepatitis C virus. ILAR J 42:152–160 [DOI] [PubMed] [Google Scholar]
- 21.Schaluder GG, Dawson GJ, Simons JN, PilotMatias TJ, Gutierrez RA, Heynen CA et al. (1995) Molecular and serologic analysis in the transmission of the GB hepatitis agents. J Med Virol 46:81–90 [DOI] [PubMed] [Google Scholar]
- 22.Simons JN, Pilot-Matias TJ, Leary TP, Dawson GJ, Desai SM, Schlauder GG et al. (1995) Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci USA 92:3401–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bukh J, Apgar CL, Yanagi M (1999) Towarda surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology 262:470–478 [DOI] [PubMed] [Google Scholar]
- 24.Lanford RE, Chavez D, Notvall L, Brasky KM (2003) Comparison of tamarins and marmot-sets as hosts for GBV-B infections and the effect of immunosuppression on duration of viremia. Virology 311:72–80 [DOI] [PubMed] [Google Scholar]
- 25.Bukh J, Apgar CL, Govindarajan S, Purcell RH (2001) Host range studies of GB virus-B hepatitis agent, the closest relative ofhepatitis C virus, in New World monkeys and chimpanzees. J Med Virol 65:694–697 [DOI] [PubMed] [Google Scholar]
- 26.Takikawa S, Engle RE, Faulk KN, Emerson SU, Purcell RH, Bukh J (2010) Molecular evolution of GB virus B hepatitis virus during acute resolving and persistent infections in experimentally infected tamarins. J Gen Virol 91:727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, Zhu S, Shuai L, Xu Y, Yin S, Bian Y et al. (2014) Infection of common marmosets with hepatitis C virus/GB virus-B chimeras. Hepatology 59:789–802 [DOI] [PubMed] [Google Scholar]
- 28.Zhu S, Li T, Liu B, Xu Y, Sun Y, Wang Y et al. (2016) Infection of common marmosets with GB virus B chimeric virus encoding the major nonstructural proteins NS2 to NS4A of hepatitis C virus. J Virol 90:8198–8211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N et al. (2012) Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol 86:6171–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaender S, Cavalleri JM, Walter S, Doerrbecker J, Campana B, Brown RJ et al. (2015) Clinical course of infection and viral tissue tropism of hepatitis C virus-like nonprimate hepaciviruses in horses. Hepatology 61:447–459 [DOI] [PubMed] [Google Scholar]
- 31.Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ et al. (2013) Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad SciUS A 110:8194–8199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corman VM, Grundhoff A, Baechlein C, Fischer N, Gmyl A, Wollny R et al. (2015) Highly divergent hepaciviruses from African cattle. J Virol 89:5876–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baechlein C, Fischer N, Grundhoff A, Alawi M, Indenbirken D, Postel A et al. (2015) Identification of a novel hepacivirus in domestic cattle from Germany. J Virol 89:7007–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauck M, Sibley SD, Lara J, Purdy MA, Khudyakov Y, Hyeroba D et al. (2013) A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild Old World primate. J Virol 87:8971–8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ et al. (2015) Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drexler JF, Corman VM, Muller MA, Lukashev AN, Gmyl A, Coutard B et al. (2013) Evidence for novel hepaciviruses in rodents. PLoS Pathog 9:e1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapoor A, Simmonds P, Scheel TK, Hjelle B, Cullen JM, Burbelo PD et al. (2013) Identification of rodent homologs of hepatitis C virus and pegiviruses. MBio 4:e00216–e00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P et al. (2014) Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. MBio 5: e01933–e01914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Billerbeck E, Wolfisberg R, Fahnoe U, Xiao JW, Quirk C, Luna JM et al. (2017) Mouse models of acute and chronic hepacivirus infection. Science 357:204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB et al. (2010) Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog 6: e1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Schaewen M, Dorner M, Hueging K, Foquet L, Gerges S, Hrebikova G et al. (2016) Expanding the host range of hepatitis C virus through viral adaptation. MBio 7: e01915–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Safran M, Kim WY, Kung AL, Horner JW, DePinho RA, Kaelin WG Jr (2003) Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cremediated recombination. Mol Imaging 2:297–302 [DOI] [PubMed] [Google Scholar]
- 43.Dorner M, Rice CM, Ploss A (2013) Study of hepatitis C virus entry in genetically humanized mice. Methods 59:249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K et al. (2011) A genetically humanized mouse model for hepatitis C virus infection. Nature 474:208–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Zhao Y, Zhang C, Chen H, Feng J, Chi X et al. (2014) Persistent hepatitis C virus infections and hepatopathological manifestations in immune-competent humanized mice. Cell Res 24:1050–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Q, von Schaewen M, Hrebikova G, Heller B, Sandmann L, Plaas M et al. (2017) Mice expressing minimally humanized CD81 and occludin genes support hepatitis C virus uptake in vivo. J Virol 91(4):e01799–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR et al. (2005) Robust hepatitis C virus infection in vitro. Proc Natl Acad SciUS A 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R et al. (1998) Binding of hepatitis C virus to CD81. Science 282:938–941 [DOI] [PubMed] [Google Scholar]
- 49.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G et al. (2002) The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J 21:5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B et al. (2007) Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805 [DOI] [PubMed] [Google Scholar]
- 51.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP et al. (2009) Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dorner M, Horwitz JA, Donovan BM, Labitt RN, Budell WC, Friling T et al. (2013) Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature 501:237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li D, von Schaewen M, Wang X, Tao W, Zhang Y, Li L et al. (2016) Altered glycosylation patterns increase immunogenicity of a subunit hepatitis C virus vaccine, inducing neutralizing antibodies which confer protection in mice. J Virol 90:10486–10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandgren EP, Palmiter RD, Heckel JL, Daugherty CC, Brinster RL, Degen JL (1991) Complete hepatic regeneration after somatic deletion of an albumin-plasminogen activator transgene. Cell 66:245–256 [DOI] [PubMed] [Google Scholar]
- 55.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A et al. (2001) Hepatitis C virus replication in mice with chimeric human livers. Nat Med 7:927–933 [DOI] [PubMed] [Google Scholar]
- 56.Tesfaye A, Stift J, Maric D, Cui Q, Dienes HP, Feinstone SM (2013) Chimeric mouse model for the infection of hepatitis B and C viruses. PLoS One 8:e77298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E et al. (2007) Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat Biotechnol 25:903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Schaewen M, Hrebikova G, Ploss A (2016) Generation of human liver chimeric mice for the study of human hepatotropic pathogens. Methods Mol Biol 1438:79–101 [DOI] [PubMed] [Google Scholar]
- 59.Hasegawa M, Kawai K, Mitsui T, Taniguchi K, Monnai M, Wakui M et al. (2011) The reconstituted ‘humanized liver’ in TK-NOG mice is mature and functional. Biochem Biophys Res Commun 405:405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kosaka K, Hiraga N, Imamura M, Yoshimi S, Murakami E, Nakahara T et al. (2013)A novel TK-NOG based humanized mouse model for the study of HBV and HCV infections. Biochem Biophys Res Commun 441:230–235 [DOI] [PubMed] [Google Scholar]
- 61.Bility MT, Zhang L, Washburn ML, Curtis TA, Kovalev GI, Su L (2012) Generation of a humanized mouse model with both human immune system and liver cells to model hepatitis C virus infection and liver immunopathogenesis. Nat Protoc 7:1608–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bility MT, Curtis A, Su L (2014) A chimeric mouse model to study immunopathogenesis of HCV infection. Methods Mol Biol 1213:379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson EM, Bial J, Tarlow B, Bial G, Jensen B, Greiner DL et al. (2014) Extensive double humanization of both liver and hematopoiesis in FRGN mice. Stem Cell Res 13:404–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gutti TL, Knibbe JS, Makarov E, Zhang J, Yannam GR, Gorantla S et al. (2014) Human hepatocytes and hematolymphoid dual reconstitution in treosulfan-conditioned uPA-NOG mice. Am J Pathol 184:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strick-Marchand H, Dusseaux M, Darche S, Huntington ND, Legrand N, Masse-Ranson G et al. (2015) A novel mouse model for stable engraftment of a human immune system and human hepatocytes. PLoS One 10:e0119820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Billerbeck E, Mommersteeg MC, Shlomai A, Xiao JW, Andrus L, Bhatta A et al. (2016) Humanized mice efficiently engrafted with fetal hepatoblasts and syngeneic immune cells develop human monocytes and NK cells. J Hepatol 65:334–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Washburn ML, Bility MT, Zhang L, Kovalev GI, Buntzman A, Frelinger JA et al. (2011) A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology 140:1334–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stoller JZ, Degenhardt KR, Huang L, Zhou DD, Lu MM, Epstein JA (2008) Cre reporter mouse expressing a nuclear localized fusion of GFP and beta-galactosidase reveals new derivatives of Pax3-expressing precursors. Genesis 46:200–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen tL, Liu CC et al. (2005) Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 70.Perkus ME, Limbach K, Paoletti E (1989) Cloning and expression of foreign genes in vaccinia virus, using a host range selection system. J Virol 63:3829–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perkus ME, Piccini A, Lipinskas BR, Paoletti E (1985) Recombinant vaccinia virus: immunization against multiple pathogens. Science 229:981–984 [DOI] [PubMed] [Google Scholar]
- 72.Youn JW, Hu YW, Tricoche N, Pfahler W, Shata MT, Dreux M et al. (2008) Evidence for protection against chronic hepatitis C virus infection in chimpanzees by immunization with replicating recombinant vaccinia virus. J Virol 82:10896–10905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J et al. (1993) Characterization ofhepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol 67:6753–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rollier CS, Paranhos-Baccala G, Verschoor EJ, Verstrepen BE, Drexhage JA, Fagrouch Z et al. (2007) Vaccine-induced early control of hepatitis C virus infection in chimpanzees fails to impact on hepatic PD-1 and chronicity. Hepatology 45:602–613 [DOI] [PubMed] [Google Scholar]
- 75.Gomez CE, Perdiguero B, Cepeda MV, Mingorance L, Garcia-Arriaza J, Vandermeeren A et al. (2013) High, broad, polyfunctional, and durable T cell immune responses induced in mice by a novel hepatitis C virus (HCV) vaccine candidate (MVA-HCV) based on modified vaccinia virus Ankara expressing the nearly full-length HCV genome. J Virol 87:7282–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abraham JD, Himoudi N, Kien F, Berland JL, Codran A, Bartosch B et al. (2004) Comparative immunogenicity analysis of modified vac-cinia Ankara vectors expressing native or modified forms of hepatitis C virus E1 and E2 glycoproteins. Vaccine 22:3917–3928 [DOI] [PubMed] [Google Scholar]
- 77.Ball JK, Tarr AW, McKeating JA (2014) The past, present and future of neutralizing antibodies for hepatitis C virus. Antivir Res 105:100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuiken C, Simmonds P (2009) Nomenclature and numbering of the hepatitis C virus. Methods Mol Biol 510:33–53 [DOI] [PubMed] [Google Scholar]
- 79.Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD Jr et al. (2009) Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J Virol 83:12473–12482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P et al. (1999) Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol 73:6235–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giang E, Dorner M, Prentoe JC, Dreux M, Evans Mj, Bukh J et al. (2012) Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci USA 109:6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johansson DX, Voisset C, Tarr AW, Aung M, Ball JK, Dubuisson J et al. (2007) Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc Natl Acad Sci U S A 104:16269–16274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keck ZY, Li TK, Xia J, Gal-Tanamy M, Olson O, Li SH et al. (2008) Definition of a conserved immunodominant domain on hepatitis C virus E2 glycoprotein by neutralizing human monoclonal antibodies. J Virol 82:6061–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J et al. (2012) Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog 8:e1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schofield DJ, Bartosch B, Shimizu YK, Allander T, Alter HJ, Emerson SU et al. (2005) Human monoclonal antibodies that react with the E2 glycoprotein of hepatitis C virus and possess neutralizing activity. Hepatology 42:1055–1062 [DOI] [PubMed] [Google Scholar]
- 86.Meunier JC, Russell RS, Goossens V, Priem S, Walter H, Depla E et al. (2008) Isolation and characterization of broadly neutralizing human monoclonal antibodies to the e1 glycoprotein of hepatitis C virus. J Virol 82:966–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Owsianka A, Tarr AW, Juttla VS, Lavillette D, Bartosch B, Cosset FL et al. (2005) Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol 79:11095–11104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stamataki Z, Coates S, Evans MJ, Wininger M, Crawford K, Dong C et al. (2007) Hepatitis C virus envelope glycoprotein immunization of rodents elicits cross-reactive neutralizing antibodies. Vaccine 25:7773–7784 [DOI] [PubMed] [Google Scholar]
- 89.Naarding MA, Falkowska E, Xiao H, Dragic T (2011) Hepatitis C virus soluble E2 in combination with QuilA and CpG ODN induces neutralizing antibodies in mice. Vaccine 29:2910–2917 [DOI] [PubMed] [Google Scholar]
- 90.Whidby J, Mateu G, Scarborough H, Demeler B, Grakoui A, Marcotrigiano J (2009) Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J Virol 83:11078–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frey SE, Houghton M, Coates S, Abrignani S, Chien D, Rosa D et al. (2010) Safety and immunogenicity of HCV E1E2 vaccine adjuvanted with MF59 administered to healthy adults. Vaccine 28:6367–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ray R, Meyer K, Banerjee A, Basu A, Coates S, Abrignani S et al. (2010) Characterization of antibodies induced by vaccination with hepatitis C virus envelope glycoproteins. J Infect Dis 202:862–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Law JL, Chen C, Wong J, Hockman D, Santer DM, Frey SE et al. (2013) A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS One 8: e59776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meyer K, Banerjee A, Frey SE, Belshe RB, Ray R (2011) A weak neutralizing antibody response to hepatitis C virus envelope glycoprotein enhances virus infection. PLoS One 6:e23699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z et al. (2008) Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med 14:25–27 [DOI] [PubMed] [Google Scholar]
- 96.Clayton RF, Owsianka A, Aitken J, Graham S, Bhella D, Patel AH (2002) Analysis of antigeicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J Virol 76:7672–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Desombere I, Fafi-Kremer S, Van Houtte F, Pessaux P, Farhoudi A, Heydmann L et al. (2016) Monoclonal anti-envelope antibody AP33 protects humanized mice against a patient-derived hepatitis C virus challenge. Hepatology 63:1120–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ruwona TB, Giang E, Nieusma T, Law M (2014) Fine mapping of murine antibody responses to immunization with a novel soluble form of hepatitis C virus envelope glycoprotein complex. J Virol 88:10459–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Balazs AB, Chen J, Hong CM, Rao DS, Yang L, Baltimore D (2011) Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481:81–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Balazs AB, Bloom JD, Hong CM, Rao DS, Baltimore D (2013) Broad protection against influenza infection by vectored immunoprophylaxis in mice. Nat Biotechnol 31:647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Balazs AB, Ouyang Y, Hong CM, Chen J, Nguyen sM, Rao DS et al. (2014) Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat Med 20:296–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deal C, Balazs AB, Espinosa DA, Zavala F, Baltimore D, Ketner G (2014) Vectored anti-body gene delivery protects against Plasmodium falciparum sporozoite challenge in mice. Proc Natl Acad Sci USA 111:12528–12532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Jong YP, Dorner M, Mommersteeg MC, Xiao JW, Balazs AB, Robbins JB et al. (2014) Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci Transl Med 6:254ra129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ji C, Liu Y, Pamulapati C, Bohini S, Fertig G, Schraeml M et al. (2015) Prevention of hepatitis C virus infection and spread in human liver chimeric mice by an anti-CD81 monoclonal antibody. Hepatology 61:1136–1144 [DOI] [PubMed] [Google Scholar]
- 105.Meuleman P, Hesselgesser J, Paulson M, Vanwolleghem T, Desombere I, Reiser H et al. (2008) Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology 48:1761–1768 [DOI] [PubMed] [Google Scholar]
- 106.Fukasawa M, Nagase S, Shirasago Y, Iida M, Yamashita M, Endo K et al. (2015) Monoclo-nal antibodies against extracellular domains of claudin-1 block hepatitis C virus infection in a mouse model. J Virol 89:4866–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mailly L, Xiao F, Lupberger J, Wilson GK, Aubert P, Duong FH et al. (2015) Clearance of persistent hepatitis C virus infection in humanized mice using a claudin-1-targeting monoclonal antibody. Nat Biotechnol 33:549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Akazawa D, Moriyama M, Yokokawa H, Omi N, Watanabe N, Date T et al. (2013) Neutralizing antibodies induced by cell culture-derived hepatitis C virus protect against infection in mice. Gastroenterology 145 (447–455):e441–e444 [DOI] [PubMed] [Google Scholar]
- 109.Vanwolleghem T, Bukh J, Meuleman P, Desombere I, Meunier JC, Alter H et al. (2008) Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology 47:1846–1855 [DOI] [PubMed] [Google Scholar]