Abstract
Excessive alcohol consumption induces intestinal dysbiosis of the gut microbiome and reduces gut epithelial integrity. This often leads to portal circulation–mediated translocation of gut-derived microbial products, such as lipopolysaccharide (LPS), to the liver, where these products engage Toll-like receptor 4 (TLR4) and initiate hepatic inflammation, which promotes alcoholic liver disease (ALD). Although the key self-destructive process of autophagy has been well-studied in hepatocytes, its role in macrophages during ALD pathogenesis remains elusive. Using WT and myeloid cell–specific autophagy-related 7 (Atg7) knockout (Atg7ΔMye) mice, we found that chronic ethanol feeding for 6 weeks plus LPS injection enhances serum alanine aminotransferase and IL-1β levels and augments hepatic C-C motif chemokine ligand 5 (CCL5) and C-X-C motif chemokine ligand 10 (CXCL10) expression in WT mice, a phenotype that was further exacerbated in Atg7ΔMye mice. Atg7ΔMye macrophages exhibited defective mitochondrial respiration and displayed elevated mitochondrial reactive oxygen species production and inflammasome activation relative to WT cells. Interestingly, compared with WT cells, Atg7ΔMye macrophages also had a drastically increased abundance and nuclear translocation of interferon regulatory factor 1 (IRF1) after LPS stimulation. Mechanistically, LPS induced co-localization of IRF1 with the autophagy adaptor p62 and the autophagosome, resulting in subsequent IRF1 degradation. However, upon p62 silencing or Atg7 deletion, IRF1 started to accumulate in autophagy-deficient macrophages and translocated into the nucleus, where it induced CCL5 and CXCL10 expression. In conclusion, macrophage autophagy protects against ALD by promoting IRF1 degradation and removal of damaged mitochondria, limiting macrophage activation and inflammation.
Keywords: alcohol, inflammasome, macrophage, autophagy, chemokine, Toll-like receptor 4 (TLR4), cytokine, alcoholic liver disease, CC-chemokine ligand 10, CC-chemokine ligand 5, dysbiosis, innate immunity
Introduction
Alcoholic liver disease (ALD)2 is a result of chronic consumption of excessive alcohol, and the clinical spectrum ranges from alcoholic fatty liver to alcoholic hepatitis, alcoholic cirrhosis, and hepatocellular carcinoma (1). 90% of heavy alcohol drinkers exhibit hepatic steatosis, and 35% of them can develop alcoholic hepatitis, a more advanced form of ALD. Among patients who develop severe alcoholic hepatitis, ∼40% of them die within 6 months after diagnosis and treatment (2). Unfortunately, corticosteroids are still used as a standard therapy for severe alcoholic hepatitis, which has not improved in the past 40 years (1, 2). A better understanding of the pathogenic mechanism of ALD is urgently needed to develop more effective therapies.
Both alcohol and its metabolite, acetaldehyde, cause hepatocyte damage, resulting in liver inflammation. Long-term overconsumption of alcohol can lead to chronic inflammation in the liver, which further enhances liver damage and dysfunction and may eventually result in liver failure (1). Therefore, it is of utmost importance to understand how chronic liver inflammation is induced as a result of excessive alcohol consumption. Previous work has shown that activation of hepatic Toll-like receptor (TLR) signaling is a key underlying mechanism of liver injury, steatosis, inflammation, and fibrosis in ALD (3). Chronic alcohol consumption is well known to cause intestinal dysbiosis and compromise intestinal barrier function, which, in turn, increases gut permeability, allowing translocation of gut microbe–derived products, such as lipopolysaccharide (LPS), to the liver through the portal circulation (4). The presence of gut-derived LPS has been shown to engage TLR4 on Kupffer cells to initiate inflammation, which drives ALD pathogenic progression (1, 2, 5, 6). Consistent with this notion, mice deficient in TLR4 or orally treated with nonabsorbable antibiotics are protected against alcohol-induced liver injury (5–7). TLR4 engagement activates two parallel signaling branches that utilize the adaptor molecules MyD88 and TIR-domain-containing adapter-inducing interferon-β (TRIF). The MyD88-dependent pathway, activated by all TLRs except for TLR3, is responsible for up-regulation of proinflammatory cytokines (e.g. TNFα, pro-IL-1β, IL-6, CXCL1, and CCL3), whereas the TRIF-mediated signaling axis (activated by TLR3 and TLR4) induces production of IFN-β, CCL5, and CXCL10 in macrophages (8). Interestingly, relative to the MyD88 signaling branch, TRIF-dependent pathway appears to have a major role in the development of ALD (6, 9). In addition to TLR4, accumulating evidence over the past decade suggests that the NLRP3 inflammasome, a key immune sensor of tissue damage, drives hepatic inflammation after chronic alcohol overconsumption, promoting ALD progression (10). Notably, TLR4 also contributes to NLRP3 inflammasome activation, as it provides a strong “priming” signal to induce de novo synthesis of pro-IL-1β as well as NLRP3 up-regulation, a crucial step required for inflammasome activation and subsequent maturation of the proinflammatory cytokine IL-1β (11).
Autophagy, an intracellular self-degradation system for cytoplasmic constituents (12, 13), is another key player that regulates the development of chronic liver diseases, including ALD. Autophagy in hepatocytes can inhibit lipid accumulation and cell death, preventing pathogenic progression of ALD and nonalcoholic fatty liver disease (NAFLD) (13, 14). Consistently, down-regulation of hepatic autophagy activity has been reported in both ALD and NAFLD (13–15). In contrast to hepatocyte autophagy, the role of macrophage autophagy in ALD pathogenesis has remained unclear. Interestingly, autophagy has been shown to inhibit NLRP3 inflammasome activation by promoting elimination of damaged mitochondria after NLRP3 activator stimulation. This ultimately prevents the generation of the NLRP3 inflammasome activators, mitochondrial ROS and oxidized mitochondrial DNA (16–18).
Based on these findings, we investigated the role of macrophage autophagy in ALD pathogenesis. We demonstrated that chronic ethanol feeding suppressed macrophage autophagy, leading to overactivation of the NLRP3 inflammasome. Moreover, decreased macrophage autophagy also results in defective autophagic degradation of IRF1, a transcription factor responsible for LPS-induced CCL5 and CXCL10 expression, exacerbating inflammation. In summary, our results provide novel insights for understanding the role of macrophage autophagy in ALD pathogenesis.
Results
Chronic ethanol consumption suppresses autophagy in liver macrophages
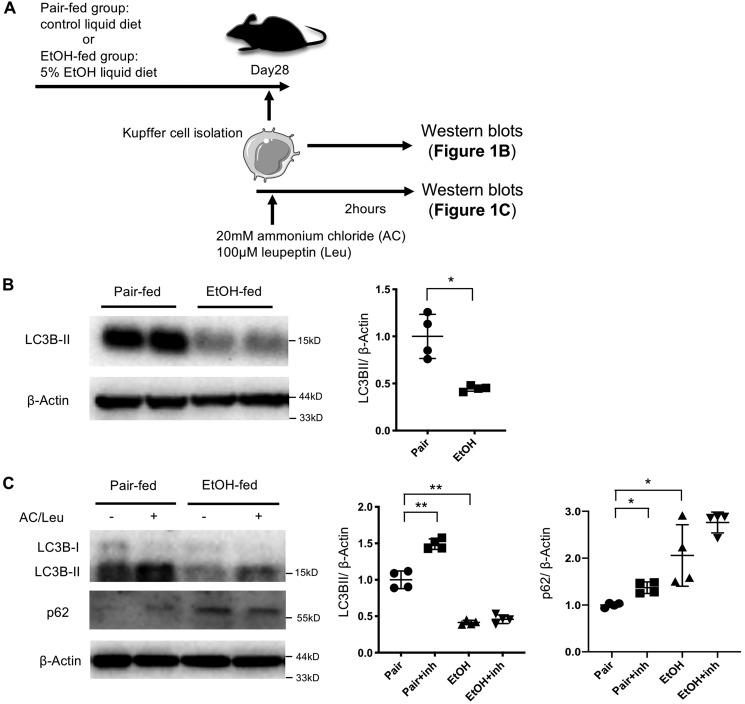
First we investigated whether autophagy is altered in liver macrophages during ALD development. LC3B is a structural protein of autophagosomes that is lipidated to attach it to the autophagosomal membrane, converting it from LC3B-I to LC3B-II during autophagy, which is commonly used as a surrogate indicator for autophagy activity (19). We found that liver macrophages from ethanol-fed mice exhibited a drastic reduction in LC3B-II abundance relative to those from control mice (Fig. 1, A and B). To determine whether autophagy flux is inhibited in liver macrophages in ALD, liver macrophages were treated with the lysosomal inhibitors ammonium chloride and leupeptin (Fig. 1A). Inhibition of lysosomal proteases increased LC3B-II and p62 levels in liver macrophages from control mice, whereas the LC3B-II expression levels in liver macrophages from ethanol-fed mice were much lower than those in liver macrophages from control mice, and inhibition of lysosomal proteases did not significantly increase either LC3B-II or p62 levels in Fig. 1, A–C. This suggests that autophagy activities are low and that autophagic flux is diminished in liver macrophages from ethanol-treated mice (19).
Figure 1.
Autophagy is suppressed in liver macrophages after chronic ethanol exposure. Liver macrophages were isolated from WT mice after 6 weeks of a paired liquid diet or a Lieber–DeCarli diet containing 5% ethanol (v/v). A, the experimental design for B and C. B, LC3B protein expression was determined by Western blotting. LC3BII levels were reduced in liver macrophages from ethanol-fed mice. C, isolated liver macrophages were treated with ammonium chloride and leupeptin (AC/Leu) for 2 h, and LC3B and p62 expression was examined by Western blotting. Similar results were obtained in four independent experiments. Representative results and their quantification are shown. *, p < 0.05; **, p < 0.01. inh, inhibitor.
Macrophage autophagy inhibits hepatic injury and steatosis induced by ethanol and LPS co-administration
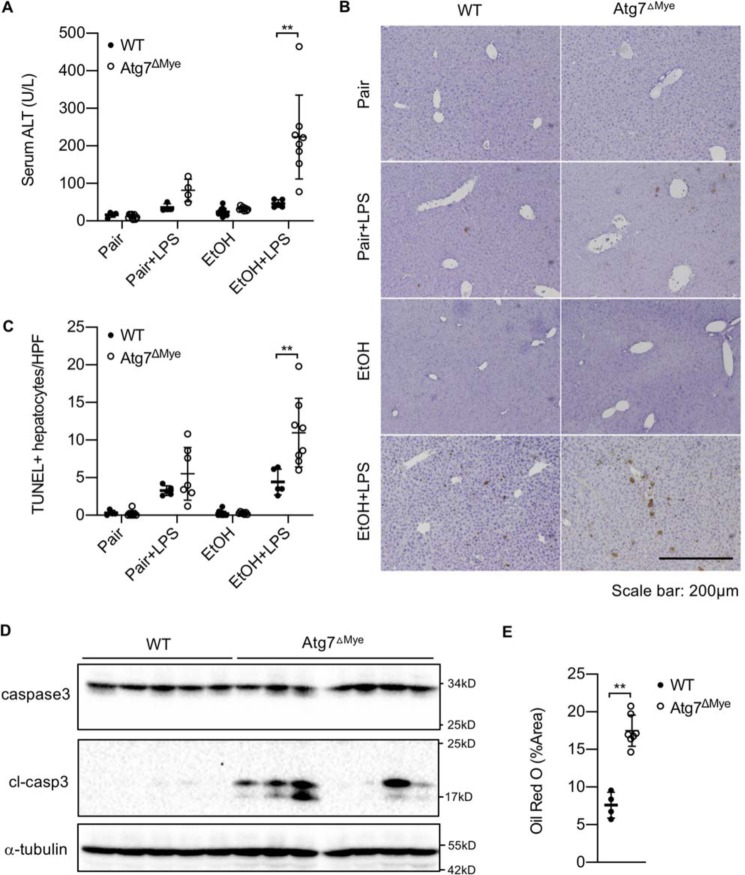
As macrophage autophagy activity is compromised in ALD, we went on to investigate the in vivo role of macrophage autophagy in alcohol-induced liver injury and steatosis using myeloid cell–specific, Atg7-deficient (Atg7ΔMye) mice that were subjected to ethanol feeding for 6 weeks. As shown in Fig. 2A, chronic ethanol feeding slightly increased serum ALT levels in WT mice, which was further exacerbated in Atg7ΔMye mice (Fig. 2A). To better mimic the pathological condition seen in ALD patients, in whom intestinal permeability is increased as a result of alcohol overconsumption, we i.p. injected a single dose of LPS, a gut-derived microbial product acting through TLR4 on liver macrophages to promote ALD development, into mice after 6 weeks of ethanol feeding. Ethanol and LPS co-administration resulted in dramatically elevated serum ALT levels in Atg7ΔMye mice compared with WT control mice (Fig. 2A). This correlated with a significant increase in the numbers of TUNEL-positive apoptotic hepatocytes as well as enhanced cell death (as measured by cleaved caspase-3 immunoblot analysis) in Atg7ΔMye mice compared with their WT littermates (Fig. 2, B–D). Additionally, hepatic steatosis was dramatically increased in Atg7ΔMye mice after ethanol and LPS co-administration (Fig. 2E). These results collectively demonstrate that macrophage autophagy inhibits liver injury and steatosis in mice after ALD is induced.
Figure 2.
Macrophage autophagy negatively regulates ethanol-induced liver injury and steatosis. A–E, WT and LysM-Cre/Atg7 flox (Atg7ΔMye) mice were treated for 6 weeks with a paired liquid diet or a Lieber–DeCarli diet containing 5% ethanol. Some mice were subsequently administered LPS (n = 5–8/group). A, serum ALT levels. B and C, apoptotic hepatocytes of liver tissues were determined by TUNEL staining. Representative pictures and quantification of TUNEL staining of liver tissues are shown. D, full-length and cleaved caspase-3 in livers from WT and Atg7ΔMye mice treated with an ethanol-containing Lieber–DeCarli diet plus LPS were determined by Western blotting. E, hepatic steatosis of WT and Atg7ΔMye mice treated with an ethanol-containing Lieber–DeCarli diet plus LPS were determined by Oil Red O staining. Quantification of Oil Red O staining is shown. **, p < 0.01.
Macrophage autophagy inhibits IL-1β production in ALD
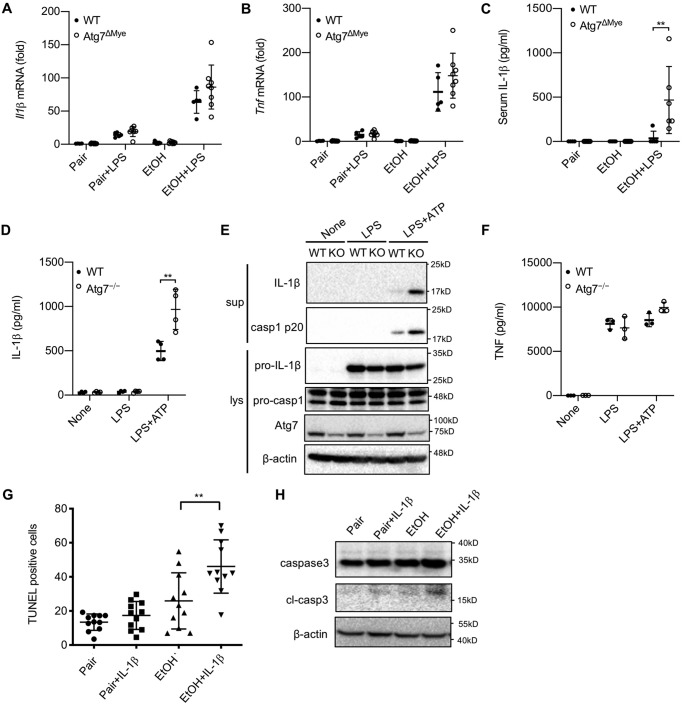
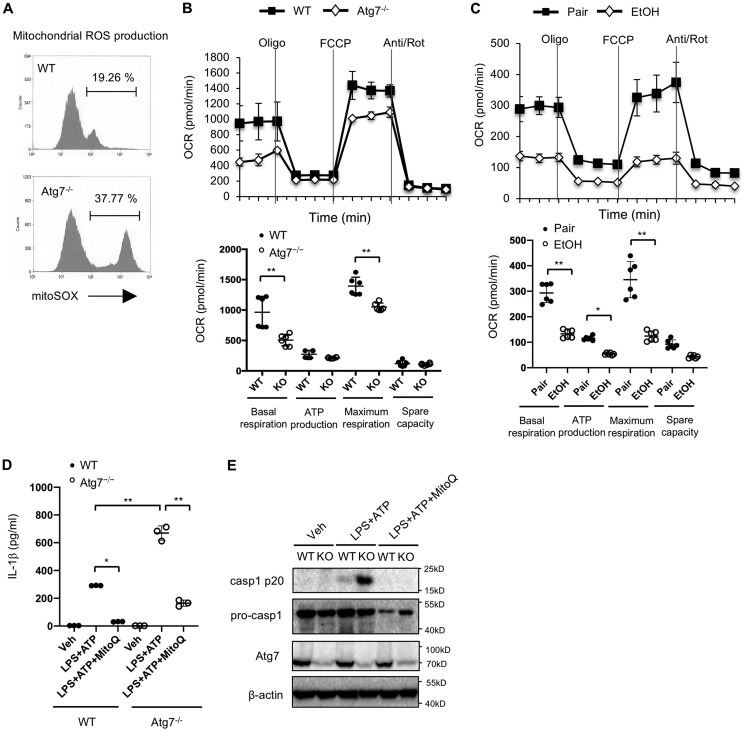
A number of previous studies have demonstrated that macrophage autophagy negatively regulates NLRP3 inflammasome activation and subsequent IL-1β maturation (16, 18, 20). We therefore quantified IL-1β expression during ALD development. Although there were no significant differences in hepatic IL-1β and TNFα mRNA levels between WT and Atg7ΔMye mice after ethanol and LPS treatment, serum IL-1β protein levels were drastically elevated in Atg7ΔMye mice after ALD was induced (Fig. 3, A–C). These data suggest that macrophage autophagy inhibits IL-1β maturation in ALD. To further confirm this notion, we measured caspase-1 cleavage as well as IL-1β production in WT and Atg7ΔMye BMDMs after LPS + ATP treatment. Indeed, deficiency in Atg7 led to much more pronounced mature IL-1β production after LPS + ATP stimulation (Fig. 3D). This further correlated with more abundant caspase-1 p20 subunits in culture supernatants from Atg7ΔMye BMDMs, although Atg7 deficiency had no effect on the expression of pro-IL-1β or TNF secretion (Fig. 3, E and F). We then investigated whether increased IL-1β by Atg7 deficiency is associated with increased death in hepatocytes from ethanol-fed Atg7ΔMye mice (Fig. 2). We examined hepatocyte apoptosis in response to recombinant IL-1β. We found that IL-1β did not increase hepatocyte death in normal hepatocytes, whereas hepatocytes isolated from ethanol diet–fed mice increased TUNEL-positive cells and cleaved caspase-3 by IL-1β treatment (Fig. 3, G and H), suggesting that ethanol feeding sensitized hepatocytes to IL-1β-induced death. Subsequently, we investigated the underlying molecular mechanism of ethanol-mediated IL-1β production. Because mitochondrial ROS are critical for NLRP3 inflammasome activation, we next compared mitochondrial ROS production in WT versus Atg7ΔMye BMDMs. As shown in Fig. 4A, Atg7 deficiency in macrophages resulted in overproduction of mitochondrial ROS (Fig. 4, A and B). These results suggest that macrophage autophagy suppresses mitochondrial ROS production, preventing NLRP3 inflammasome activation as well as IL-1β maturation. We then examined whether Atg7 deficiency and ethanol can compromise mitochondrial function in liver macrophages by measuring the oxygen consumption rate (OCR). As shown in Fig. 4, B and C, liver macrophages from Atg7ΔMye mice and ethanol-fed WT mice had a lower OCR relative to control diet–fed mice. These results suggest that autophagy suppression by chronic ethanol feeding could be associated with compromised mitochondrial function. Last, we examined whether mitochondrial ROS contributes to IL-1β production. MitoQ is a selective inhibitor of mitochondrial ROS (16). MitoQ treatment suppressed IL-1β production and caspase-1 activation (Fig. 4, D and E), suggesting that increased mitochondrial ROS in Atg7ΔMye BMDMs promotes inflammasome activation, producing IL-1β. Taken together, these results suggest that chronic ethanol consumption reduces macrophage autophagy, causing an accumulation of dysfunctional mitochondria and ROS in liver macrophages, promoting NLRP3 inflammasome activation and IL-1β production.
Figure 3.
Macrophage autophagy negatively regulates activation of caspase-1 and IL-1β maturation. A and B, WT and LysM-Cre/Atg7 flox (Atg7ΔMye) mice were treated with a paired liquid diet or a Lieber–DeCarli diet containing 5% ethanol for 6 weeks. Some mice were subsequently administered LPS (n = 5–8/group). A and B, hepatic mRNA expression of IL-1β (A) and TNF (B) was determined by quantitative real-time PCR. C, serum IL-1β levels were determined by ELISA. C and D, WT and Atg7−/− (KO) BMDMs were treated with LPS (100 ng/ml) for 24 h with or without ATP (2 mm). D, IL-1β levels in the supernatant were measured by ELISA. E, IL-1β and caspase-1 in the supernatant (sup) and cell lysate (lys) were determined by Western blotting. casp, caspase. F, TNF mRNA expression was determined by quantitative real-time PCR. G and H, TUNEL staining (G) and Western blotting (H) for caspase-3 using primary hepatocytes isolated from mice fed a paired liquid diet or a Lieber-DeCarli diet containing 5% ethanol. Cells were treated with IL-1β for 24 h. Similar results were obtained in three independent experiments. Representative results are shown. **, p < 0.01.
Figure 4.
Macrophage autophagy negatively regulates activation of caspase-1 and IL-1β maturation through mitochondrial ROS production. A, WT and Atg7−/− (KO) BMDMs were treated with LPS (100 ng/ml) for 24 h. Mitochondrial ROS was determined by MitoSOX. B, OCRs of WT and Atg7−/− BMDMs were determined by Seahorse bioanalyzer. FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; Rot, rotenone. C, OCR in liver macrophages was assessed. The OCR was suppressed in ethanol-treated liver macrophages. D and E, WT and Atg7−/− BMDMs were treated with LPS (100 ng/ml) and ATP (2 mm) with or without MitoQ (5 μm). D, IL-1β levels in the supernatant were measured by ELISA. Veh, vehicle. E, Western blots for caspase-1 (casp1). Similar results were obtained in three independent experiments. Representative results are shown. *, p < 0.05; **, p < 0.01.
Macrophage autophagy prevents CCL5 and CXCL10 production in ALD
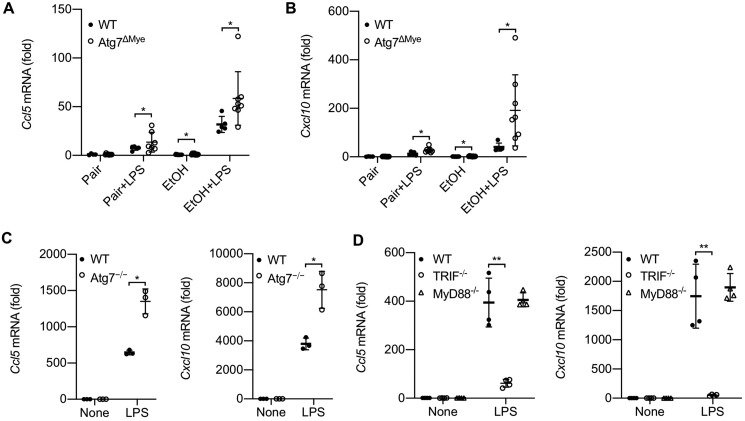
As autophagy maintains a cellular healthy status by concurrently monitoring a number of cellular processes, we explored additional mechanism(s) by which macrophage autophagy could prevent alcohol-induced liver injury and steatosis. In addition to NLRP3 inflammasome overaction, we observed that Atg7ΔMye mice also exhibited increased production of CCL5 and CXCL10 after LPS or chronic ethanol feeding (Fig. 5, A and B). Notably, chronic ethanol feeding plus LPS injection induced a further elevation of CCL5 and CXCL10 levels in Atg7ΔMye mice relative to their WT littermates (Fig. 5, A and B). In line with this notion, Atg7ΔMye BMDMs, compared with WT cells, produced more CCL5 and CXCL10 after LPS stimulation (Fig. 5C). CCL5 and CXCL10 induction is mediated by the TLR4-TRIF pathway (8). Indeed, macrophages deficient in TRIF, but not MyD88, abolished LPS-induced expression of CCL5 and CXCL10 (Fig. 5D). These results together suggest that, in addition to antagonizing NLRP3 inflammasome activation, macrophage autophagy also inhibits CCL5 and CXCL10 production and may thereby further prevent alcohol-mediated liver injury, inflammation, and steatosis.
Figure 5.
LPS-induced, TRIF-dependent CCL5 and CXCL10 expression is negatively regulated by autophagy in macrophages. A and B, WT and LysM-Cre/Atg7 flox (Atg7ΔMye) mice were treated for 6 weeks with a paired liquid diet or a Lieber–DeCarli diet containing 5% ethanol. Some mice subsequently received LPS (n = 5–8/group). Hepatic mRNA expression of CCL5 and CXCL10 was determined by quantitative real-time PCR. C, WT and Atg7ΔMye BMDMs were treated with LPS (100 ng/ml) for 4 h. CCL5 and CXCL10 mRNA expression was determined by quantitative real-time PCR. D, WT, TRIF−/−, and MyD88−/− BMDMs were treated with LPS (100 ng/ml) for 4 h. CCL5 and CXCL10 mRNA expression was determined by quantitative real-time PCR. Similar results were obtained in three independent experiments. Representative results are shown. *, p < 0.05; **, p < 0.01.
Macrophage autophagy promotes IRF1 degradation
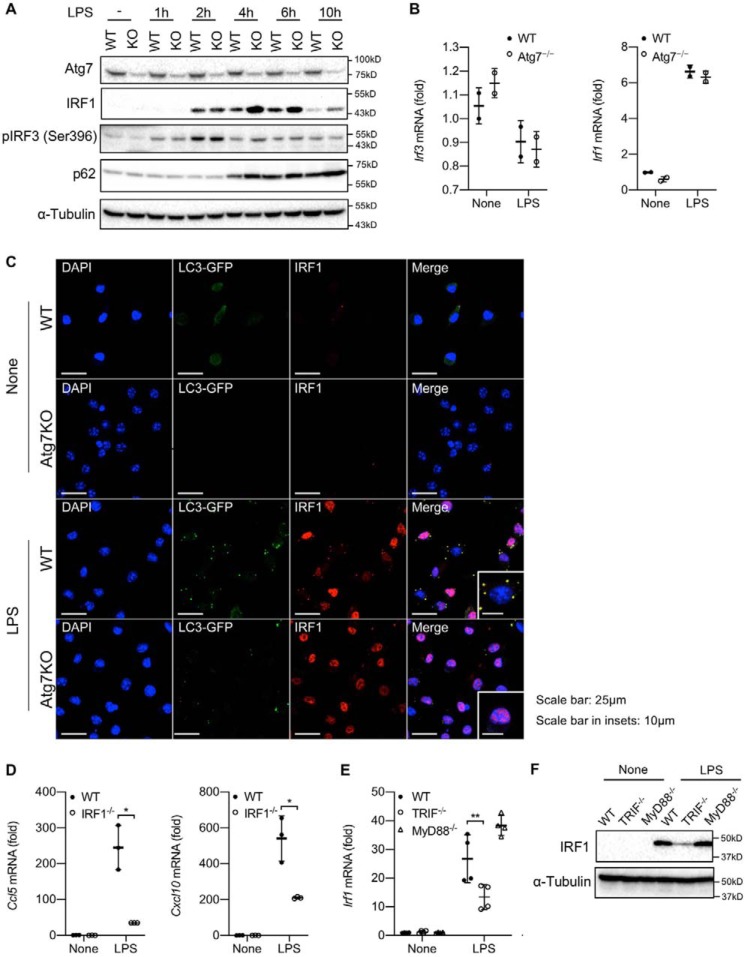
To further delineate how macrophage autophagy inhibits CCL5 and CXCL10 production, we examined whether any of the TLR4-TRIF pathway downstream signaling molecules may be regulated by autophagy. Engagement of the TLR4–TRIF–dependent pathway is known to activate the transcription factor IRF3. However, we did not observe any apparent differences in the levels of IRF3 mRNA or phosphorylated IRF3 protein in Atg7-deficient macrophages relative to WT cells over 10 h after LPS treatment (Fig. 6, A and B). In addition to IRF3, it has been reported that IRF1 is required for IL-1β–induced CCL5 and CXCL10 transcription (21). Interestingly, IRF1 protein but not mRNA abundance was dramatically increased in Atg7-deficient macrophages relative to WT cells as early as 4 h after LPS stimulation (Fig. 6, A and B). We therefore examined whether autophagy participates in posttranslational regulation of IRF1 in macrophages. By taking advantage of BMDMs from LC3B-GFP transgenic mice, we visualized autophagosome formation by microscopy. Upon LPS stimulation, IRF1 expression started to increase, and some IRF1 proteins were already translocated into the nucleus (Fig. 6C). Notably, a significant fraction of IRF1 was retained in the cytosol, where it co-localized with LC3-GFP–labeled autophagosomes (Fig. 6C). However, in Atg7-deficient LC3B-GFP transgenic macrophages where autophagosome formation was disrupted, the majority of IRF1 proteins were already translocated into the nucleus (Fig. 6C). Together, these results clearly demonstrate that autophagy inhibits IRF1 protein degradation and prevents its nuclear translocation, inhibiting IRF1-dependent transcription of downstream genes. Indeed, we found that IRF1 is required for LPS-induced CCL5 and CXCL10 production (Fig. 6D) and further confirmed that induction of IRF1 by LPS is largely dependent on the TRIF-dependent pathway (Fig. 6, E and F). Taken together, these results suggest that macrophage autophagy prevents LPS-induced CCL5 and CXCL10 expression by promoting the degradation of IRF1.
Figure 6.
Macrophage autophagy regulates cellular levels of IRF1. A–C, WT and Atg7−/− (KO) BMDMs were treated with LPS (100 ng/ml) up to 10 h (A) and for 4 h (B and C). A, Atg7, IRF1, phospho-IRF-3, total IRF3, and p62 expression was determined by Western blotting. B, IRF3 and IRF1 mRNA expression was determined by quantitative real-time PCR. C, BMDMs from WT and Atg7−/− (KO) LC3B-GFP transgenic mice were used for immunofluorescence. Green, red, and blue represent LC3B, IRF1, and nucleus, respectively. Yellow indicates co-localization of LC3B and IRF1. D, WT and IRF1−/− BMDMs were treated with LPS (100 ng/ml) for 4 h. CCL5 and CXCL10 mRNA expression was determined by quantitative real-time PCR. E and F, WT, TRIF−/−, and MyD88−/− BMDMs were treated with LPS (100 ng/ml) for 4 h. IRF1 mRNA and protein expression was determined by quantitative real-time PCR (E) and Western blotting (F), respectively. Similar results were obtained in three independent experiments. Representative results are shown. *, p < 0.05; **, p < 0.01.
Autophagic degradation of IRF1 requires p62/SQSTM1
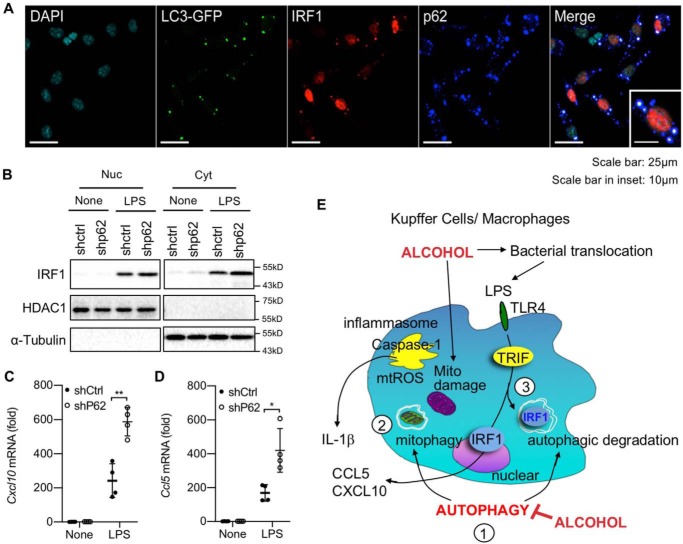
As p62/SQSTM1 is a key adaptor protein that connects the autophagic cargo with autophagosome (14), we next examined whether p62 participates in autophagy-mediated IRF1 degradation. Indeed, IRF1 was co-localized with both LC3B autophagosome and p62 aggregates after LPS stimulation (Fig. 7A). Moreover, p62-deficient macrophages (shp62 immortalized BMDMs) had increased IRF1 in both the cytoplasmic and nuclear fractions compared with control macrophages after LPS stimulation (Fig. 7B). Consistently, LPS-induced CCL5 and CXCL10 production was also significantly higher in p62-silenced macrophages relative to control cells (Fig. 7, C and D).
Figure 7.
p62/SQSTM1 regulates cellular levels of IRF1 through autophagy in macrophages. A, BMDMs from WT-LC3B-GFP transgenic mice were treated with LPS (100 ng/ml) for 4 h. Immunofluorescence for IRF1 and p62 was performed. Cyan, green, red, and blue represent nucleus, LC3B, IRF1, and p62. White indicates co-localization of LC3B, IRF1, and p62. DAPI, 4′,6-diamidino-2-phenylindole. B–D, control and p62-silenced BMDMs were treated with LPS (100 ng/ml) for 4 h. B, nuclear (Nuc) and cytosolic (Cyt) fractions were separated. IRF1, HDAC1, and α-tubulin expression was determined by Western blotting. C and D, CCL5 (C) and CXCL10 (D) mRNA expression was determined by quantitative real-time PCR. Similar results were obtained in three independent experiments. Representative results are shown. *, p < 0.05; **, p < 0.01. E, proposed mechanisms. 1, chronic alcohol consumption inhibits macrophage autophagy. 2, suppressed mitophagy causes accumulation of damaged mitochondria, which is associated with mitochondrial dysfunction, and produces mitochondrial ROS, resulting in activation of the inflammasome and IL-1β. 3, autophagy regulates cellular IRF1 levels through autophagic degradation. TLR4–TRIF–mediated IRF1 expression regulates CCL5 and CXCL10 production. Produced IL-1β, CCL5, and CXCL10 promote the development of liver injury and inflammation in ALD.
Discussion
Alcohol consumption is known to alter hepatic autophagy. Acute ethanol exposure enhances autophagy activity, whereas chronic ethanol consumption inhibits it (22). It is generally accepted that autophagy in hepatocytes prevents hepatocyte death and liver steatosis in ALD (13–15). However, whether and how macrophage autophagy plays a role in ALD remains unknown. In this study, we demonstrate that macrophage autophagy, which is compromised as a result of chronic ethanol consumption, negatively regulates alcohol-induced liver injury, steatosis, and hepatic inflammation and thus inhibits ALD development. Mechanistically, macrophage autophagy drives the removal of damaged mitochondria and thereby limits mitochondrial ROS production as well as NLRP3 inflammasome activation and subsequent inflammation. Additionally, macrophage autophagy also promotes p62-mediated degradation of IRF1 to inhibit IRF1-dependent transcription of CCL5 and CXCL10, two important chemokines involved in liver inflammation (see the schematic summary in Fig. 7E).
Chronic ethanol treatment has been shown to sensitize liver macrophages (23), which can enhance the response to LPS. The underlying mechanisms proposed include reduced expression of IL-1 receptor-associated kinase (IRAK)-M, an endogenous inhibitor of TLR/IL-1 receptor signaling, and heme oxygenase-1 (HO-1), an antioxidant (24, 25). In this study, we provided evidence that autophagy inhibition in liver macrophages is an additional mechanism by which chronic ethanol consumption leads to ALD development. Importantly, we demonstrated that defective macrophage autophagy leads to accumulation of damaged mitochondria as well as IRF1 protein, which drives ALD development. Damaged mitochondria are normally eliminated by mitophagy, a selective autophagy for mitochondria. However, chronic ethanol exposure compromised macrophage autophagy, causing defective clearance of damaged mitochondria, which enhances mitochondrial ROS production as well as mitochondrial DNA oxidization, which participate in induction of NLRP3 inflammasome activation and liver inflammation.
The most interesting discovery of this study is that macrophage autophagy promotes degradation of IRF1. Prevention of autophagic degradation of IRF1 as a result of Atg7 or p62 deficiency increased IRF1 nuclear translocation, augmenting CCL5 and CXCL10 expression. This pathway is mainly regulated by a TLR4–TRIF–dependent pathway, which has been reported to be crucial for ALD development (21, 26). Intriguingly, our research team has recently demonstrated that IRF1 contributes to NLRP3 inflammasome activation by inducing expression of CMPK2, which promotes mitochondrial DNA synthesis (27). We speculate that increased IRF1 levels as a result of defective macrophage autophagy could also promote NLRP3 inflammasome activation through CMPK2.
Our results are also consistent with previous reports showing that macrophage autophagy inhibits NLRP3 inflammasome activation through suppression of ROS production (17, 18, 28), preventing the development of diseases associated with immune overactivation. For instance, in an acute liver injury model induced by galactosamine plus LPS, macrophage autophagy prevented liver injury by inhibiting IL-1β production (16, 20). Indeed, our data demonstrated that suppression of increased mitochondria ROS production by macrophage autophagy deficiency reduced caspase-1 activation and IL-1β production. In addition to the link between autophagy and the inflammasome, macrophage autophagy has also been shown to regulate M1 versus M2 macrophage polarization. In mice with autophagy-deficient macrophages, liver macrophages tend to polarize to the M1 phenotype, which promotes liver injury and inflammation in NAFLD (29), although the underlying mechanisms remain unknown. In summary, this study has not only demonstrated that chronic ethanol consumption inhibits macrophage autophagy but also provided novel mechanistic insights into how defective macrophage autophagy promotes ALD development, which may provide insights for designing new therapies to treat and/or prevent alcoholic liver diseases.
Experimental procedures
Animal Experiments
C57BL/6 WT mice, TRIF-mutant lps2 mice, IRF1-deficient mice, and LysM-Cre transgenic mice were purchased from The Jackson Laboratory (Bar Harbor, ME). MyD88-deficient mice were originally generated by Dr. Akira (Osaka University, Suita, Japan), and Atg7flox/flox mice were originally generated by Dr. Komatsu (Niigata University, Niigata, Japan) (30). Atg7ΔMye mice were generated by crossing Atg7flox/flox mice with LysM-Cre transgenic mice. Atg7flox/flox mice without the Cre transgene were used as WT littermate controls for Atg7ΔMye mice. All mice, including WT mice, were bred in the University of California San Diego and Cedars-Sinai Medical Center vivaria. All genetically modified mice were backcrossed at least 10 generations onto the C57BL/6 background and displayed a similar hepatic phenotype as WT mice with standard laboratory chow. Female mice were used for in vivo experiments.
For induction of mouse ALD, mice were fed a control liquid diet ad libitum for the first 5 days as an acclimatization step and subsequently fed a Lieber–DeCarli diet (Bio-Serv, Frenchtown, NJ) containing 5% ethanol (v/v) for 6 weeks. Some of these mice were then challenged with LPS (1 mg/kg intraperitoneally) and sacrificed 8 h after LPS administration.
All studies were in accordance with National Institutes of Health recommendations outlined in the Guide for the Care and Use of Laboratory Animals. All animal experimental protocols were approved by the University of California San Diego and Cedars-Sinai Medical Center Institutional Animal Care and Use Committee.
Histological analysis
Mouse liver tissues were collected, fixed in 10% neutral buffered formalin solution for 48 h, routinely processed, and then embedded in paraffin. Tissue sections (4 μm) were prepared using a microtome and placed on glass slides. TUNEL staining was performed. TUNEL-positive cells were counted on at least 8 random fields per slide and expressed as cells per high-power field.
RNA isolation and quantitative real-time PCR analysis
RNA was extracted from mouse liver tissues and cells using TRIzol (Life Technologies) and a column kit (NucleoSpin®, Clontech, Mountain View, CA) and treated with DNase I (Promega, Madison, WI). Extracted RNA was converted to complementary DNA using a reverse transcription kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Quantitative real-time PCR was then performed using a CFX96 Real Time System (Bio-Rad) using SYBR Green I as a double-stranded DNA–specific binding dye. After the reaction was completed, specificity was verified by melting curve analysis. Quantification was performed by comparing the Ct values of each sample with normalization to 18S RNA. Sequences of primers are summarized in Table S1.
Cell culture experiments
Liver macrophages and primary hepatocytes were isolated from mice fed a paired liquid diet or a Lieber-DeCarli diet containing 5% ethanol as described previously (31). Liver macrophages were cultured in RPMI 1640 medium (Gibco, Life Technologies). To examine autophagic flux, cells were treated with 20 mm ammonium chloride and 100 μm leupeptin (Sigma) for 2 h. Primary bone marrow–derived macrophages (BMDMs) were generated by culturing mouse bone marrow cells in the presence of 20% (v/v) L929 conditional medium for 7 days as described previously (16). LPS (100 ng/ml, Sigma) and ATP (2 mm, Sigma) were used to stimulate liver macrophages or BMDMs. Cells were also treated with MitoQ (5 μm, MedKoo, Morrisville, NC). p62-silenced BMDMs were generated as described previously (16). Primary hepatocytes from mice fed a paired liquid diet or a Lieber-DeCarli diet containing 5% ethanol were treated with recombinant IL-1β (10 ng/ml) for 24 h, followed by TUNEL staining and western blotting for caspase-3.
Measurement for ALT and IL-1β
Serum alanine aminotransferase (ALT) levels were measured by Infinity ALT reagent (Thermo, Waltham, MA). The levels of IL-1β in the serum and cell supernatant were analyzed by ELISA kit (R&D Systems, Minneapolis, MN).
TG extraction and measurement
Hepatic triglycerides (TGs) were extracted as described previously (32). TG content was measured using a triglyceride measurement kit (Pionte Scientific, Canton, MI) according to the manufacturer's instructions.
Western blots
Protein extracts were electrophoresed and then blotted. Blots were incubated with antibody for LC3B, IL-1β, caspase-3, cleaved caspase-3, Atg7, IRF1, IRF3, phospho-IRF3 (Cell Signaling Technology, Danvers, MA), mouse caspase-1 p20 (Adipogen, San Diego, CA), p62 (ProGen, Heidelberg, Germany), tubulin (Santa Cruz Biotechnology, Dallas, TX), and β-actin (Sigma).
Mitochondrial ROS detection
Mitochondrial ROS were measured using MitoSOX (Thermo). Briefly, macrophages were treated with LPS (100 ng/ml) for 24 h, washed twice with PBS, loaded with 4 μm MitoSOX for 20 min, and washed twice with PBS. Fluorescence intensity was determined using a FACSCanto flow cytometer (BD Biosciences).
Measurement of mitochondrial oxygen consumption
OCRs were measured using an XF24 Extracellular Flux Seahorse Bioanalyzer (Agilent, Santa Clara, CA). For the XF24 assay, cells were equilibrated with DMEM lacking bicarbonate at 37 °C for 1 h in an incubator without CO2. Oligomycin, which blocks phosphorylation of ADP to ATP, was utilized to prevent mitochondrial respiration and to provide basal O2 consumption during the assay. Carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) was used as an uncoupling agent to allow maximal O2 consumption under a given condition. Rotenone and antimycin were employed as a mitochondrial respiratory chain complex I inhibitor and a complex III inhibitor, respectively.
Immunofluorescent staining and confocal microscopy
BMDM were treated with or without LPS (100 ng/ml) for 2 h. After the treatment, the cells were washed twice with sterile PBS, fixed with 4% paraformaldehyde, permeabilized with 0.01% Triton X-100, and blocked in 2% BSA and 1% donkey serum. The cells were then incubated overnight with primary antibodies, including anti-p62 (ProGen), and anti-IRF1 (Cell Signaling Technology). Secondary fluorescent antibodies (Alexa 488, 594, or 647; Life Technologies) were added for 1 h, and 4′,6-diamidino-2-phenylindole (DAPI) was used for nuclear counterstaining. Samples were imaged using a SP5 confocal microscope (Leica) 24 h after mounting.
Statistical analysis
Statistical significance was assessed using GraphPad Prism 8 software (GraphPad Software, Inc.). Differences between the two groups were compared using a two-tailed unpaired Student's t test. Differences between multiple groups were compared using one-way analysis of variance, followed by Tukey's post hoc analysis. p < 0.05 was considered significant.
Author contributions
S. L. and E. S. conceptualization; S. L., S. Y. K., R. U., Y. S. R., and H. M. data curation; S. L., S. Y. K., R. U., Y. S. R., H. M., and E. S. formal analysis; S. L. and E. S. investigation; S. L., Z. Z., and R. A. G. methodology; S. L., Z. Z., R. A. G., and E. S. writing-review and editing; E. S. supervision; E. S. funding acquisition; E. S. writing-original draft.
Supplementary Material
This study was supported by National Institute on Alcohol Abuse and Alcoholism Grants R01AA027036 and R21AA025841 (to E. S.); NIDDK, National Institutes of Health Grant R01DK085252 (to E. S.); an American Liver Foundation Congressman John Joseph Moakley postdoctoral research fellowship (to Y. S. R.); National Research Foundation of Korea Grant 2017R1C1B2004423 (to Y. S. R.); and a Cedars-Sinai Medical Center Winnick Research Award (to E. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1.
- ALD
- alcoholic liver disease
- TLR
- Toll-like receptor
- LPS
- lipopolysaccharide
- TNF
- tumor necrosis factor
- NAFLD
- non-alcoholic fatty liver disease
- ROS
- reactive oxygen species
- ALT
- alanine aminotransferase
- BMDM
- bone marrow–derived macrophage
- OCR
- oxygen consumption rate
- TG
- triglyceride
- TRIF
- TIR-domain-containing adapter-inducing interferon-β.
References
- 1. Gao B., and Bataller R. (2011) Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 141, 1572–1585 10.1053/j.gastro.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lucey M. R., Mathurin P., and Morgan T. R. (2009) Alcoholic hepatitis. N. Engl. J. Med. 360, 2758–2769 10.1056/NEJMra0805786 [DOI] [PubMed] [Google Scholar]
- 3. Szabo G., Petrasek J., and Bala S. (2012) Innate immunity and alcoholic liver disease. Dig. Dis. 30, 55–60 10.1159/000341126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seki E., and Schnabl B. (2012) Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J. Physiol. 590, 447–458 10.1113/jphysiol.2011.219691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Inokuchi S., Tsukamoto H., Park E., Liu Z. X., Brenner D. A., and Seki E. (2011) Toll-like receptor 4 mediates alcohol-induced steatohepatitis through bone marrow-derived and endogenous liver cells in mice. Alcohol Clin. Exp. Res. 35, 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hritz I., Mandrekar P., Velayudham A., Catalano D., Dolganiuc A., Kodys K., Kurt-Jones E., and Szabo G. (2008) The critical role of Toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology 48, 1224–1231 10.1002/hep.22470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Uesugi T., Froh M., Arteel G. E., Bradford B. U., and Thurman R. G. (2001) Toll-like receptor 4 is involved in the mechanism of early alcohol-induced liver injury in mice. Hepatology 34, 101–108 10.1053/jhep.2001.25350 [DOI] [PubMed] [Google Scholar]
- 8. Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., and Akira S. (2003) Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643 10.1126/science.1087262 [DOI] [PubMed] [Google Scholar]
- 9. Petrasek J., Dolganiuc A., Csak T., Nath B., Hritz I., Kodys K., Catalano D., Kurt-Jones E., Mandrekar P., and Szabo G. (2011) Interferon regulatory factor 3 and type I interferons are protective in alcoholic liver injury in mice by way of crosstalk of parenchymal and myeloid cells. Hepatology 53, 649–660 10.1002/hep.24059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Petrasek J., Bala S., Csak T., Lippai D., Kodys K., Menashy V., Barrieau M., Min S. Y., Kurt-Jones E. A., and Szabo G. (2012) IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice. J. Clin. Invest. 122, 3476–3489 10.1172/JCI60777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsutsui H., Imamura M., Fujimoto J., and Nakanishi K. (2010) The TLR4/TRIF-mediated activation of NLRP3 inflammasome underlies endotoxin-induced liver injury in mice. Gastroenterol. Res. Pract. 2010, 641865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yin X. M., Ding W. X., and Gao W. (2008) Autophagy in the liver. Hepatology 47, 1773–1785 10.1002/hep.22146 [DOI] [PubMed] [Google Scholar]
- 13. Czaja M. J., Ding W. X., Donohue T. M. Jr., Friedman S. L., Kim J. S., Komatsu M., Lemasters J. J., Lemoine A., Lin J. D., Ou J. H., Perlmutter D. H., Randall G., Ray R. B., Tsung A., and Yin X. M. (2013) Functions of autophagy in normal and diseased liver. Autophagy 9, 1131–1158 10.4161/auto.25063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y., Wang S., Ni H. M., Huang H., and Ding W. X. (2014) Autophagy in alcohol-induced multiorgan injury: mechanisms and potential therapeutic targets. Biomed Res. Int. 2014, 498491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. von Haefen C., Sifringer M., Menk M., and Spies C. D. (2011) Ethanol enhances susceptibility to apoptotic cell death via down-regulation of autophagy-related proteins. Alcohol Clin. Exp. Res. 35, 1381–1391 [DOI] [PubMed] [Google Scholar]
- 16. Zhong Z., Umemura A., Sanchez-Lopez E., Liang S., Shalapour S., Wong J., He F., Boassa D., Perkins G., Ali S. R., McGeough M. D., Ellisman M. H., Seki E., Gustafsson A. B., Hoffman H. M., et al. (2016) NF-κB restricts inflammasome activation via elimination of damaged mitochondria. Cell 164, 896–910 10.1016/j.cell.2015.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi C. S., Shenderov K., Huang N. N., Kabat J., Abu-Asab M., Fitzgerald K. A., Sher A., and Kehrl J. H. (2012) Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol 13, 255–263 10.1038/ni.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saitoh T., Fujita N., Jang M. H., Uematsu S., Yang B. G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., Tanaka K., Kawai T., Tsujimura T., Takeuchi O., Yoshimori T., and Akira S. (2008) Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456, 264–268 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- 19. Mizushima N., Yoshimori T., and Levine B. (2010) Methods in mammalian autophagy research. Cell 140, 313–326 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ilyas G., Zhao E., Liu K., Lin Y., Tesfa L., Tanaka K. E., and Czaja M. J. (2016) Macrophage autophagy limits acute toxic liver injury in mice through down regulation of interleukin-1β. J. Hepatol. 64, 118–127 10.1016/j.jhep.2015.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harikumar K. B., Yester J. W., Surace M. J., Oyeniran C., Price M. M., Huang W. C., Hait N. C., Allegood J. C., Yamada A., Kong X., Lazear H. M., Bhardwaj R., Takabe K., Diamond M. S., Luo C., et al. (2014) K63-linked polyubiquitination of transcription factor IRF1 is essential for IL-1-induced production of chemokines CXCL10 and CCL5. Nat. Immunol. 15, 231–238 10.1038/ni.2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thomes P. G., Trambly C. S., Fox H. S., Tuma D. J., and Donohue T. M. Jr. (2015) Acute and chronic ethanol administration differentially modulate hepatic autophagy and transcription factor EB. Alcohol Clin. Exp. Res. 39, 2354–2363 10.1111/acer.12904 [DOI] [PubMed] [Google Scholar]
- 23. Enomoto N., Schemmer P., Ikejima K., Takei Y., Sato N., Brenner D. A., and Thurman R. G. (2001) Long-term alcohol exposure changes sensitivity of rat Kupffer cells to lipopolysaccharide. Alcohol Clin. Exp. Res. 25, 1360–1367 10.1111/j.1530-0277.2001.tb02359.x [DOI] [PubMed] [Google Scholar]
- 24. Mandrekar P., Bala S., Catalano D., Kodys K., and Szabo G. (2009) The opposite effects of acute and chronic alcohol on lipopolysaccharide-induced inflammation are linked to IRAK-M in human monocytes. J. Immunol. 183, 1320–1327 10.4049/jimmunol.0803206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mandal P., Pritchard M. T., and Nagy L. E. (2010) Anti-inflammatory pathways and alcoholic liver disease: role of an adiponectin/interleukin-10/heme oxygenase-1 pathway. World J. Gastroenterol. 16, 1330–1336 10.3748/wjg.v16.i11.1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang L., and Seki E. (2012) Toll-like receptors in liver fibrosis: cellular crosstalk and mechanisms. Front. Physiol. 3, 138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhong Z., Liang S., Sanchez-Lopez E., He F., Shalapour S., Lin X. J., Wong J., Ding S., Seki E., Schnabl B., Hevener A. L., Greenberg H. B., Kisseleva T., and Karin M. (2018) New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560, 198–203 10.1038/s41586-018-0372-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakahira K., Haspel J. A., Rathinam V. A., Lee S. J., Dolinay T., Lam H. C., Englert J. A., Rabinovitch M., Cernadas M., Kim H. P., Fitzgerald K. A., Ryter S. W., and Choi A. M. (2011) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu K., Zhao E., Ilyas G., Lalazar G., Lin Y., Haseeb M., Tanaka K. E., and Czaja M. J. (2015) Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy 11, 271–284 10.1080/15548627.2015.1009787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., and Tanaka K. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- 31. Roh Y. S., Zhang B., Loomba R., and Seki E. (2015) TLR2 and TLR9 contribute to alcohol-mediated liver injury through induction of CXCL1 and neutrophil infiltration. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G30–41 10.1152/ajpgi.00031.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miura K., Yang L., van Rooijen N., Brenner D. A., Ohnishi H., and Seki E. (2013) Toll-like receptor 2 and palmitic acid cooperatively contribute to the development of nonalcoholic steatohepatitis through inflammasome activation in mice. Hepatology 57, 577–589 10.1002/hep.26081 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.