Figure 9.
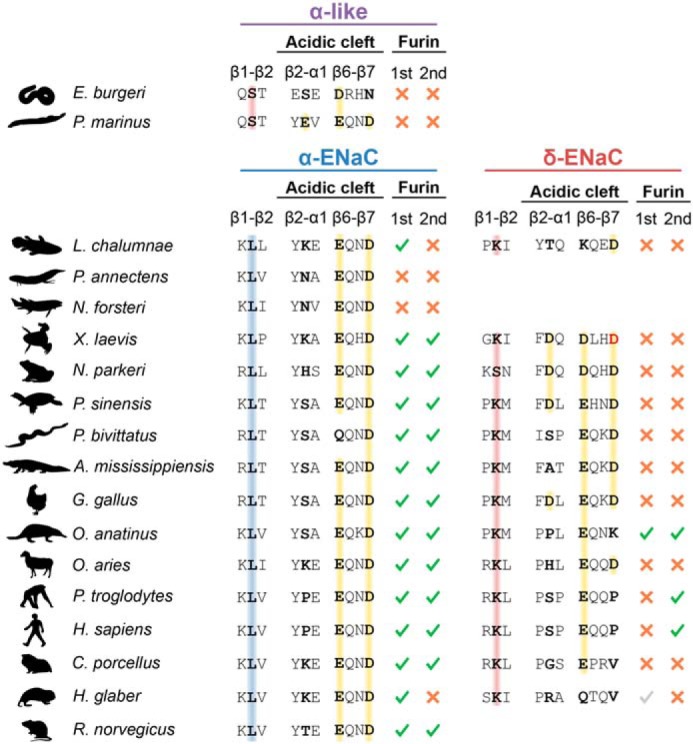
Conservation of motifs involved in pH-sensitivity and proteolytic maturation of α- and δ-ENaC orthologs. Multiple sequence alignment inferred from amino acid sequences of α- and δ-ENaC orthologs (Table S1) showing sites involved in proton-mediated activation of X. laevis δβγ-ENaC that have been identified in this study (β1–β2, β2–α1, and β1–β2 linker). Note that only one α-like subunit ortholog has been identified in hagfish Eptatretus burgeri and lamprey Petromyzon marinus, which therefore are displayed in a separate group. Rats do not have a functional gene for δ-ENaC. Colored backgrounds illustrate the presence of residues with hydrophobic (blue) or polar (red) side chains in the β1–β2 linker or negatively-charged residues (yellow) in the acidic cleft. The Asp-296 position, which is essential for Na+ coordination in Xenopus δβγ-ENaC, is highlighted in red. Conservation of the minimal consensus sequence for proteolytic maturation of ENaC subunits by furin (sequence RXXR) is indicated by green check marks, and red crosses represent the absence of this consensus sequence. Gray checkmarks indicate the presence of an RXXR motif dislocated from the conserved location of the respective furin cleavage site.
