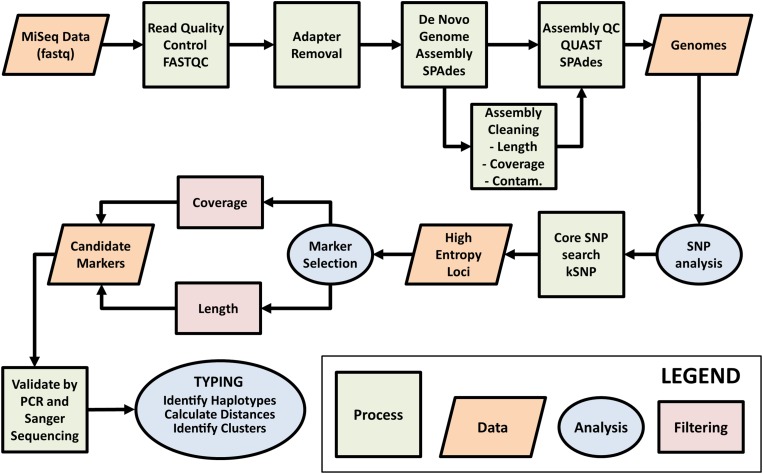
Fig. 1.
Workflow for selection of Cyclospora cayetanensis typing markers. Raw genome sequence data generated on the Illumina MiSeq platform were assessed for quality using FASTQC. AdaptorRemoval v2.1.7 (Schubert et al., 2016) was used to remove adaptor sequences from reads and to merge overlapping paired reads into consensus sequences. SPAades v3.9.0 (Bankevich et al., 2012) was used to de novo assemble the reads. During the assembly cleaning process, contigs derived from contaminating (Contam.) prokaryotic human gut flora were removed using BBMap (http://sourceforge.net/projects/bbmap/). The assemblies were assessed for quality using QUAST v4.3 (Gurevich et al., 2013) before and after the cleaning phase. Contigs with 60 times coverage, greater than or equal to 3000 base pairs (bp) long and with coding regions identified using GeneMark-ES v4.33 (Borodovsky and Lomsadze, 2011), were retained as part of the core genome. Single nucleotide polymorphisms (SNPs) were detected across the core genome assemblies using kSNP v3.021 (Gardner et al., 2015) and this information was used to identify high-entropy genomic loci. Genomic regions containing high confidence SNPs (i.e. those SNPs within genomic regions of the highest coverage) occurring within SNP-dense regions (i.e. where several informative SNPs exist within a genomic region of less than 1 kilobase pair in size), were identified as candidate typing markers for validation by PCR amplification and Sanger sequencing. The markers with the highest amplification and sequencing success rate were considered ideal candidates for C. cayetanensis typing, and were PCR amplified and sequenced from stool specimens provided by a diverse range of patients. The resulting sequences were then subjected to typing.