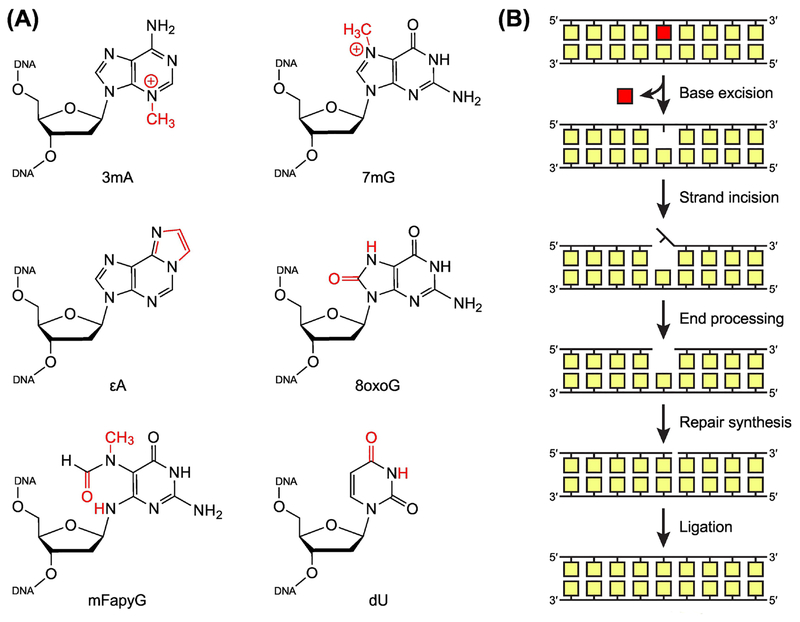
Figure 1. Base Excision Repair of Damaged Nucleobases.
(A) Common DNA lesions resulting from alkylation, oxidation, and deamination of nucleobases. 3mA, 3-methyl-2’-deoxyadenosine; 7mG, 7-methyl-2’-deoxyguanosine; 8oxoG, 8-oxo-2’-deoxyguanosine; εA, 1,N6-etheno-2’-deoxyadenosine; mFapyG, methyl derivative of N6-(2’-deoxyribosyl)-2,6-diamino-4-oxo-5-formamidopyrimidine (FapyG); dU, 2’-deoxyuridine. (B) General steps in the base excision repair (BER) pathway. BER is initiated by lesion-specific DNA glycosylases, which remove damaged nucleobases to create an apurinic/apyrimidinic (AP) site. An AP endonuclease (or a bifunctional DNA glycosylase) then incises the modified strand, producing a single-strand break. As necessary, the break is processed by one of several enzymes to create a gap with a 3’-hydroxyl group and a 5’-phosphoryl group. A DNA polymerase fills the gap with new DNA, and a DNA ligase seals the strand to complete repair. In eukaryotes, if strand incision is performed by an AP endonuclease, repair synthesis occurs before end processing, displacing the AP site.