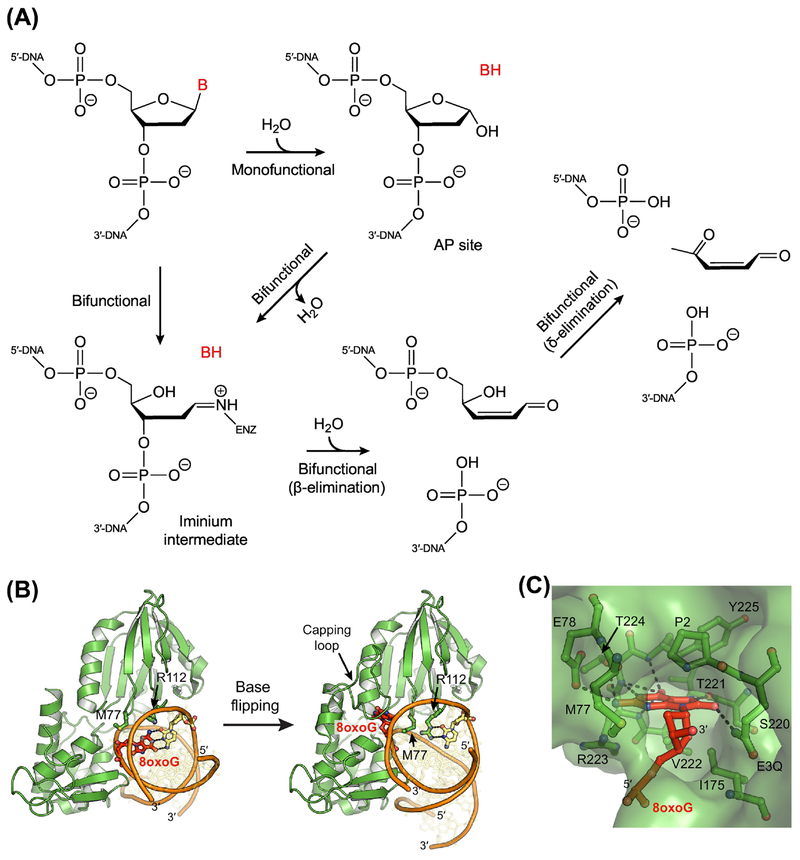
Figure 2. Initiation of Base Excision Repair by DNA Glycosylases.
(A) Base excision and strand incision reactions performed by monofunctional and bifunctional DNA glycosylases. Monofunctional enzymes catalyze only base excision, wherein the glycosidic bond between the nucleobase and the phosphodeoxyribose backbone is hydrolyzed, removing the nucleobase and creating an AP site. Bifunctional enzymes catalyze both base excision and strand incision (lyase activity). During removal of the nucleobase, most bifunctional glycosylases form an iminium intermediate, which covalently links the protein and the DNA. Some bifunctional enzymes, however, initially hydrolyze the glycosidic bond to create an AP site before then converting the AP site to an iminium intermediate. Following base excision, all bifunctional DNA glycosylases incise the strand on the 3’-side of the AP site (β-elimination), generating a single-strand break with a 3’-phospho-α,β-unsaturated aldehyde (PUA) group and a 5’-phosphoryl group. Some bifunctional enzymes also subsequently incise the strand on the 5’-side of the PUA (δ-elimination), leaving a 3’-phosphate, which must also be removed prior to repair synthesis, requiring the phosphatase activity of a separate enzyme. Alternatively, if β-elimination occurs following strand incision by an AP endonuclease, a gap is generated with the 3’-hydroxyl and 5’-phosphoryl groups necessary for synthesis and ligation. (B) X-ray crystal structures of the bifunctional DNA glycosylase Fpg (green) bound to DNA (orange and yellow) containing an 8oxoG lesion (red; PDB ID: 3GO8, 3GPY) [82]. After base flipping, Met77, Arg112, and Phe114 (not shown) fill the void in the duplex. (C) An extrahelical 8oxoG lesion in the nucleobase binding pocket of Fpg. Hydrogen-bonding interactions are indicated with dashed lines.