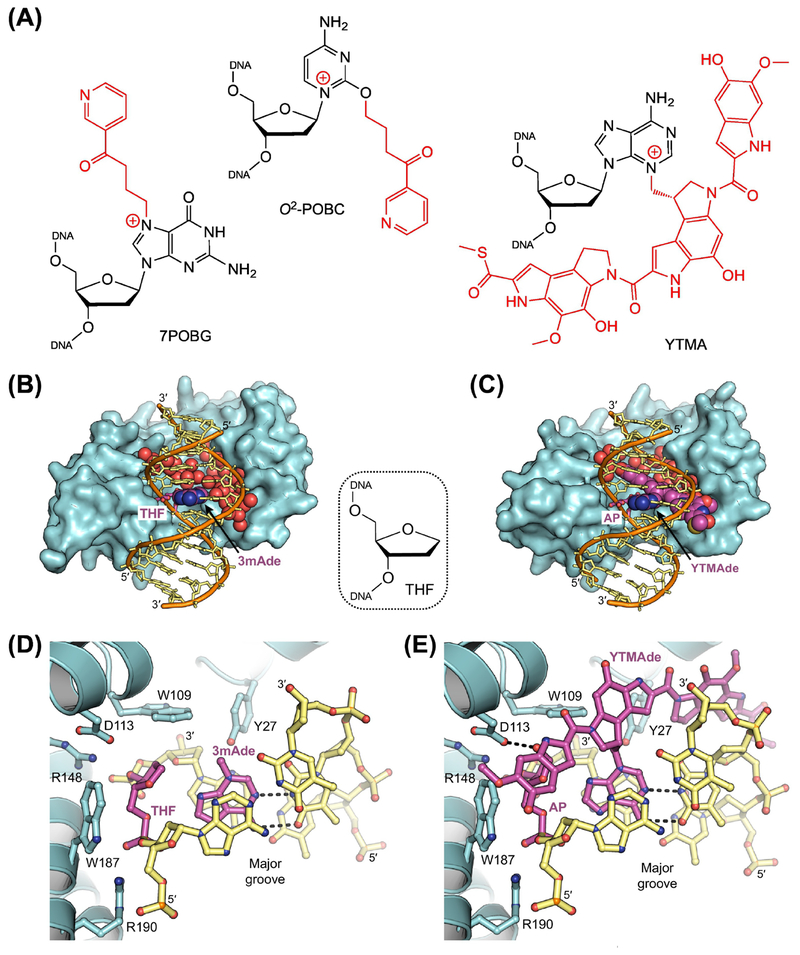
Figure 3. Removal of Bulky Adducts by AlkD.
(A) Bulky adducts excised by AlkD. 7POBG, 7-pyridyloxobutyl-2’-deoxyguanosine; O2-POBC, O2-pyridyloxobutyl-2’-deoxycytidine; YTMA, 3-yatakemycinyl-2’-deoxyadenosine. AlkD eliminates bulky lesions with modifications located in either the major (7POBG) or the minor (O2-POBC and YTMA) groove. (B,D) X-ray crystal structure of AlkD (cyan) in complex with free 3-methyladenine (3mAde) nucleobase (purple) and DNA (orange and yellow) containing a tetrahydrofuran (THF) spacer (purple) to mimic an AP site (PDB ID: 5CLE) [29]. Water molecules located in the large cavity between the protein and the DNA are depicted as red spheres. (C,E) X-ray crystal structure of AlkD in complex with an excised yatakeymycinyladenine (YTMAde) nucleobase (purple) and DNA containing an AP site (purple; PDB ID: 5UUF) [26]. Hydrogen-bonding interactions are indicated with dashed lines. Unlike DNA glycosylases that use a traditional base-flipping mechanism, the catalytic residues (Trp109, Asp113, and Trp187) present in AlkD are located on the protein surface, not recessed in a nucleobase binding pocket. Without a catalytic requirement for base flipping, the lack of protein-DNA contacts in the major groove and the large solvent-filled cavity between the protein and the minor groove allow AlkD to recognize and excise nucleobases with bulky modifications at any position.