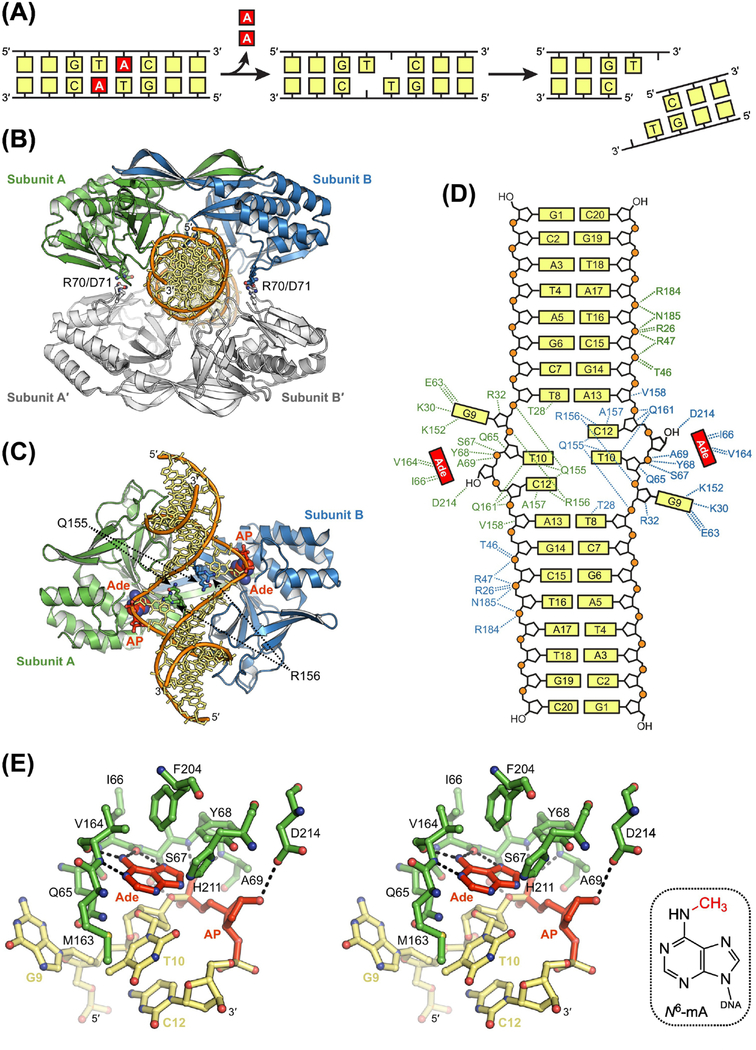
Figure 6. Sequence-Specific Excision of Adenine by R.Pabl.
(A) Double-strand break resulting from dual base excision and subsequent incision of AP sites on opposing strands. (B) X-ray crystal structure of tetrameric R.PabI (blue, green, and white) bound to non-specific DNA (orange and yellow; PDB ID: 5IFF) [94]. Four salt bridges formed by Arg70 and Asp71 are the only interactions between the two R.PabI dimers in the tetrameric search complex. (C) Crystal structure of dimeric R.PabI bound to specific DNA after dual excisions of adenine (Ade) nucleobase (red) to create AP sites (red) on opposing strands (PDB ID: 3WAZ) [90]. Binding of the recognition sequence induces a transition to a dimeric excision complex in which the GTAC base pairs are pulled apart and the void created in the duplex is stabilized by insertion of Gln155 and Arg156. (D) Hydrogen-bonding interactions in the dimeric product complex. DNA binding residues are colored according to protein subunit. R.PabI forms 13 sequence-specific and 21 non-sequence-specific hydrogen-bonding interactions with each strand in the palindromic product. (E) Recognition of Ade in the active site of R.PabI (stereodiagram). Hydrogen-bonding interactions are indicated with dashed lines. N6-methyl-2’-deoxyadenosine, N6-mA. Hydrogen bonds between Ade and backbone atoms in Ile66 and Val164 select for an N7-protonated substrate to catalyze excision. Sequence-specific methylation of Ade by M.PabI introduces steric clashes with these same backbone atoms to prevent catalytically productive binding and excision of N6-mA.